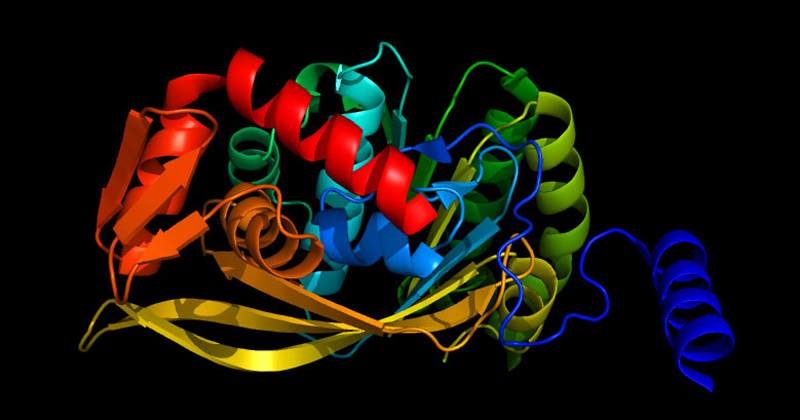
Biological life requires thousands of different protein families, about 70% of which are ‘globular’ proteins, each with a 3-dimensional shape that is unique to each family of proteins. An example is shown in the picture at the top of this post. This 3D shape is necessary for a particular biological function and is determined by the sequence of the different amino acids that make up that protein. In other words, it is not biology that determines the shape, but physics. Sequences that produce stable, functional 3D structures are so rare that scientists today do not attempt to find them using random sequence libraries. Instead, they use information they have
obtained from reverse-engineering biological proteins to intelligently design artificial proteins.
Indeed, our 21st century supercomputers are not powerful enough to crunch the variables and locate novel 3D structures. Nonetheless, a foundational prediction of neo-Darwinian theory is that a ploddingly slow evolutionary process consisting of genetic drift, mutations, insertions and deletions must be able to ‘find’ not just one, but thousands of sequences pre-determined by physics that will have different stable, functional 3D structures. So how does this falsifiable prediction hold up when tested against real data? As ought to be the case in science, I have made available my program so that you can run your own data and verify for yourself the kinds of probabilities these protein families represent. More.
Readers? Sensible responses wanted. (It’s getting so Darwin’s tenured trolls have nothing to offer but sneers, persecution, and—in the case of those afflicted with religiosity—Jesus-hollers in response.)
Follow UD News at Twitter!