Rarely does the public catch a glimpse of how Darwinists actually behave toward colleagues who disagree with their view of biological origins. Thus, as a public service, I’m presenting here a correspondence, initiated by Darwinists and unsolicited by our side, that provides readers of this blog with such a glimpse. Briefly, a Johns Hopkins biologist named David Levin sent an unsolicited and wonderfully insulting letter to Michael Behe (the entire letter is given toward the bottom of this post). Levin also attached a pdf of a Nature article (see the very bottom of this post). As it is, Levin copied Ken Miller, Richard Dawkins, and the usual suspects. Ken Miller, thinking that Levin had a slam-dunk against ID, then suggested to Levin that he also send me what he had sent Behe (presumably to crush my spirits). Here, then, is the exchange. To trace the chronology, you’ll need to start from the bottom and work your way up. I post Mike Behe’s response to Levin with Mike’s permission. After Behe’s response and my second response to Levin, we never heard from him again.
—– Original Message —–
Date: Thu, 22 Feb 2007 23:36:44 -0600
To: “David E. Levin”
From: “William A. Dembski”
Subject: Re: Fw: Evolution of a biochemical pathway by gene duplication and specialization
Cc: richard.dawkins AT zoo.ox.ac.uk, robison AT nucleus.harvard, aorr AT mail.rochester, rdoolittle AT ucsd, Kenneth_Miller AT Brown, lziska AT asrr.arsusda, Lisa West
Dear Dr. Levin,
I receive many unsolicited emails asking me to comment on how the theory of intelligent design deals with this or that objection to it. You are asking me to respond to an informal letter that you wrote to Michael Behe sketching out some worries you have about his notion of irreducible complexity. Let me suggest you write up your thoughts in a formal article and submit them to a peer-reviewed publication. Once it’s accepted, I’ll be happy to look at it more closely and offer comment. As it is, Michael Behe was gracious enough to send you some comment on your letter (I’ve pasted his comments below for continuity), though he appears much less impressed with your work than you are.
In your note to me below you write: “you seem incapable or unwilling to discuss the data or the inescapable conclusion that emerges from them.” Actually I’m quite willing. If you would like me to speak at your campus on the topic of intelligent design and address your data, I can put you in touch with my speakers bureau.
Best wishes,
Bill Dembski
—– Original Message —–
At 09:26 AM 2/22/2007, David E. Levin wrote:
Dr. Dembski,
You seem to have missed the point of my letter to Behe. It was not to bring to his attention the Kellis et al. paper. That was merely the starting point, the prerequisite understanding from which my work followed. It did not escape my notice that you had nothing whatever to say about my demonstration of how a real biochemical pathway has evolved to a more complex state. Perhaps all the biochemistry and genetics is beyond you.
Still, your criticism of the Kellis et al. paper was telling. Lets see, you asserted that it is now three years old, as though its age as some bearing on the validity of its conclusions. You clued into the phrases computational algorithm and statistical analysis as though such things invalidate any conclusions the authors might derive. These guys sequenced and assembled the genome of a species and aligned its eight chromosomes with the 16 chromosomes of another species. Yes, they used computers and statistics to assist them in their analysis. Its a 10 megabase eukaryotic genome! You sort of need computers and statistics to crunch all that information.
Whats important here is to look at the remarkable picture that emerges from this work. A species ancestral to the bakers’ yeast underwent a whole-genome duplication, followed by loss of most of the duplicated genes. This is how bakers’ yeast arrived at current genomic organization. As I said before, there is no other way to interpret these data. But you seem incapable or unwilling to discuss the data or the inescapable conclusion that emerges from them. I am taken aback by the extreme level of intellectual dishonesty that pervades the intelligent design circle. Your tactic is always to deflect and misdirect. Never mind the data, its old, or it uses statistics, or it presupposes common descent. Why are you so afraid of the data? Its as though you creationists have closed your eyes, covered your ears with your hands and are muttering to yourselves My mind is made up, dont confuse me with facts.
As for your assertion that nonteleological evolutionary mechanisms are not sufficient to drive the evolutionary process, I have provided an excellent example of precisely how this happens. Deal with it!
David E. Levin, Ph. D.
Professor
Dept. of Biochem. & Molec. Biol.
Johns Hopkins Bloomberg School of Public Health
615 N. Wolfe St.
Baltimore, MD 21205
Ph. (410) 955-snip
fax (410) 955-snip
—– Original Message —–
At 01:26 PM 2/22/2007, Michael Behe wrote:
Hello, Professor Levin, nice to meet you. Well, I see that even though you work in Baltimore, you’ve managed to avoid acquiring any Southern charm. Most folks consider it rude to send insulting, unsolicited mail to people you’ve never met, even if you don’t like their views. I hope at least you are polite toward people who agree with you.
Thanks for sending me the brief report on your work. Clearly you are excited about it, so I hope you don’t mind that I find it unimpressive even if your interpretation of events is correct. Here’s how I see your scenario: Roughly a hundred million years ago the ancestor of S. cerevisiae had a well-regulated, multicomponent pathway, including a prodigy protein, Mpk1, that had several activities. That complex pathway is taken by you for granted, as an unexplained starting point. Then the genome duplicated. In one of the duplicated copies of the prodigy protein a point mutation caused it to lose one of its pre-existing abilities. In the other copy of the prodigy protein, although it hasn’t happened yet in nature, you have in your own lab demonstrated that, by golly, a simple mutation can cause the other pre-existing ability to be lost too. I’m afraid I find all of that unsurprising. It has been known for quite a long time that mutations can inactivate protein functions.
The single gain of function in your whole story is the new binding site for Rlm1. That, however, is a comparably modest change; since the consensus binding sequence for MADS-box proteins is about ten nucleotides, with considerable redundancy, such a sequence would be expected to occur by chance perhaps every ten kilobases or so, and either to have been present in some segment of the population at the time of the genome duplication, or to be produced by point mutation very shortly thereafter. Such sites are thought to be gained and lost continually. At the very best then, assuming that modern Mpk1 eventually does lose the ability to activate SBF, according to your own scenario we are left with yeast that does pretty much the same thing with two very similar pathways that its ancestor did with one. And that meager (potential) result required enormous evolutionary resources: a hundred million years, whole genome duplication, and huge numbers of yeast likely many orders of magnitude more than the numbers of a vertebrate species that would be available in a similar span of time.
Frankly, I’m puzzled why that is supposed to be an example of the power of Darwinian processes. Id be happy to cite it myself as an illustration of genome drift within tight limits set by severe constraints. The trivial changes the scenario involves would be expected to have been available in the yeast population a very short time after the initial genome duplication event. Yet here we are twiddling our thumbs, tens of millions of years later, still waiting for the scenario to complete itself. This suggests to me that your scenario is overlooking many complicating factors, such straightforward issues as whether genome duplications or gain/loss of regulatory binding sites or loss of protein function even in a duplicated copy are deleterious, and whether there are useful functions close by existing functions. Such questions plague any simplistic Darwinian scenario, including the ones you cite that were proposed for the blood clotting cascade, but it seems few people are willing to take the difficulties seriously.
I wish you well with your work, Professor Levin. But please don’t write to me again unless you can restrain your childish sneers.
Sincerely,
Mike Behe
P.S. – I apologize for bothering all the people who were copied by Professor Levin on his original email.
—– Original Message —–
From: William A. Dembski
To: David E. Levin
Cc: richard.dawkins AT zoo.ox.ac.uk ; robison AT nucleus.harvard ; aorr AT mail.rochester ; rdoolittle AT ucsd ; Kenneth_Miller AT Brown ; lziska AT asrr.arsusda ; Michael J. Behe
Sent: Wednesday, February 21, 2007 10:29 PM
Subject: Re: Fw: Evolution of a biochemical pathway by gene duplication and specialization
Dear Dr. Levin,
Thanks for following Ken Miller’s advice and emailing me that NATURE article as well as your letter to Mike Behe (I’ve appended both below so that the people I’m copying here know what we’re talking about).
I’m sorry you feel you got the worse end of the bargain trading Behe’s book for Dawkins’s latest. I don’t own THE GOD DELUSION, but I happen to have a spare copy of Ken Miller’s FINDING DARWIN’S GOD, which I would be happy to exchange with you for Behe’s. You can send it to P.O. Box 22397, Fort Worth, Texas 76122-0397 and I’ll be sure to pop Miller’s in the mail to you.
I’m not sure what the excitement is with Kellis et al.’s NATURE article for which you sent me the pdf. The paper is now three years old and was circulated on a list that I moderate on 4.8.04. It struck me as forgettable at the time. Looking at it more closely now, the paper seems based on a statistical analysis — the authors call it a “computation algorithm” — of the gene locations, etc. Unless the authors justified their algorithm in a way that does not presuppose common descent, it’s another case of circular reasoning.
In any case, the issue for ID is not common descent (many of us in the ID community are happy to accept it or at least use it as a working hypothesis). The issue is whether nonteleological evolutionary mechanisms are sufficient to drive the evolutionary process. Ken Miller, as usual, mischaracterized the application of my mathematical work to biology. I don’t argue that evolution cannot produce biological information. Rather, I argue that nonteleological evolutionary mechanisms cannot account for a certain form of biological information, to wit, specified complexity (note that not every biological system need exhibit this form of information).
I used to think that Ken Miller was deliberately mischaracterizing my views. I’ve now come to suspect that age and inertia have simply caught up with him.
Again, thanks for your email. And please keep me in mind for preprints and articles that are a bit more current.
Best wishes,
Bill Dembski
—– Original Message —–
From: “David E. Levin”
To:
Subject: Fw: Evolution of a biochemical pathway by gene duplication and specialization
Date: Wed, 21 Feb 2007 13:38:56 -0500
Dr. Dembski,
Ken Miller asked that I forward to you this piece of work from my laboratory, sent initially to Michael Behe.
David Levin
—– Original Message —–
From: David E. Levin
To: mjb1 AT lehigh
Cc: richard.dawkins AT zoo.ox.ac.uk ; robison AT nucleus.harvard ; aorr AT mail.rochester ; rdoolittle AT ucsd ; Kenneth_Miller AT Brown ; lziska AT asrr.arsusda ; Lisa West ; David Levin
Sent: Wednesday, February 21, 2007 11:27 AM
Subject: Evolution of a biochemical pathway by gene duplication and specialization
Dr. Behe,
I thought you might be interested in the attached evidence from my laboratory for the evolution of a biochemical pathway to an increased level of complexity through gene duplication and subsequent specialization. A surprisingly small number of genetic alterations were required for this transition.
David Levin
David E. Levin, Ph. D.
Professor
Dept. of Biochem. & Molec. Biol.
Johns Hopkins Bloomberg School of Public Health
615 N. Wolfe St.
Baltimore, MD 21205
Ph. (410) 955-9825
fax (410) 955-2926
—– Original Message —–
From: Kenneth Miller
To: David E. Levin
Sent: Wednesday, February 21, 2007 12:57 PM
Subject: Re: Evolution of a biochemical pathway by gene duplication and specialization
Dear David,
Thanks for this wonderful paper and splendid piece of scientific work. May I suggest that you send a copy to my “friend” William Dembski? He argues that evolution cannot produce new biological information, and he might find your work to be “interesting” reading! E-mail: wdembski AT designinference or dembski AT discovery.
Sincerely,
Ken
PS: I’d send it myself…. but you’re the one who deserves the satisfaction! 😉
[[Oh, Ken. You’re so wonderfully clever! –WmAD]]
—
Kenneth R. Miller
Professor of Biology
Box G-B5
Brown University
Providence, RI 02912
http://research.brown/research/profile.php?id=1100924768&r=1
http://www.millerandlevine/km/index.html
=====letter from Levin to Behe =====
Dr. Michael H. Behe
Department of Biological Sciences
Lehigh University
Bethlehem, PA 18015
February 21, 2007
Dear Dr. Behe;
Having recently read Richard Dawkins latest book (The God Delusion), I traded it to a creationist friend for your book on intelligent design. It is now clear to me that my friend got the better end of the bargain. Many of the weaknesses inherent to your argument that irreducible complexity of a structure or a biochemical pathway precludes its having evolved in a gradual stepwise manner were evident directly from some of your various examples. However, just a modest review of the scientific literature beyond your presentation revealed a pattern of omission of critical information that utterly decimates every one of them. Back in 1996, many critics of your book charitably chalked this up to your ignorance of specific details. However, in the face of an enormous volume of published criticism and detailed evidence countering your claims (not to mention the Dover trial), your new afterward was striking for its glib assertion that there is very little of the original text I would change if I wrote it today. So much for ignorance as an excuse. Rather, your attitude seems disingenuous at best.
Nowhere is your need to misdirect more evident than in your flippant dismissal of Keith Robisons clearly presented argument that enzymatic cascades can develop gradually. Robison described in his review of your book a plausible step-by-step mechanism through which irreducible complexity of a biochemical pathway could be achieved through gene duplication and subsequent specialization. By this route, components arise or are recruited that are initially advantageous to pathway function, but later evolve to a state in which they are essential to its function. Your response, I encourage him to develop the argument rigorously and submit it to a refereed journal for publication. If he does so, he will be the first., fails to address the plausibility of this mechanism, which is the issue at hand. After all, you assert on page 90 and similarly elsewhere that In order to claim that a system developed gradually by a Darwinian mechanism a person must show that the function of the system could have been formed by numerous successive, slight modifications. The same argument was advanced by Allen Orr in his review of your book. You dismiss Orrs eloquent, but hypothetical treatment of the subject in your afterward as fanciful and an abstract argument [that] says nothing about the concrete sorts of examples I cited, or that commonly occur in the cell. Again, you fail to address the plausibility of the mechanism.
In the unlikely event that you are actually interested in evidence, I have provided in the following section a real example of a biochemical pathway whose evolution to a more complex state can be clearly delineated. Indeed, this pathway has acquired three of four genetic alterations necessary to add a new level of irreducible complexity. The fourth is a simple alteration that we have made in the lab, which certainly could arise spontaneously. It is clear that this example fulfills your requirements for the plausible evolution of an irreducibly complex biochemical pathway.
The story starts with the necessary recognition that an ancestral species to the bakers yeast Saccharomyces cerevisiae at some point underwent a whole-genome duplication (WGD). This event was followed by loss of most of the duplicated genes. In case you are not prepared to accept this fact, let me fill in the details. An ancestral WGD was first suggested about a decade ago when the yeast genome sequence was completed. The bakers yeast genome is unusual in that it possesses a high number (12%) of closely paralogous genes. It was observed that the 16 yeast chromosomes could be roughly paired up using closely related genes as landmarks. However, the genes between paralogs were unrelated, suggesting that if the ancestral species experienced a WGD, most of the duplicate genes were lost by deletion. This argument by itself is, of course, not very strong. Incontrovertible evidence came two years ago, with the genome sequence of another fungal species, Kluyvermyces waltii (Kellis et al., 2004 Nature 428:617), which diverged from the Saccharomyces lineage at a time prior to the presumptive WGD event. Kluyvermyces waltii possesses eight chromosomes, each of which can be aligned with the two related copies of the smaller bakers yeast chromosomes. From this alignment, it is easy to see which genes were deleted from which copy of the duplicated yeast chromosomes, thereby confirming that the Saccharomyces genome resulted from a WGD, followed by deletion of most of the paralogous genes. There is, quite frankly, no other way to interpret these data.
The Kellis et al. analysis of paralogous Saccharomyces genes and Kluyvermyces genes revealed that in many cases, one copy was evolutionarily restrained to carry out the ancestral function, whereas the other was free to diverge at an accelerated rate. Such is the case for the gene pair that is the subject of our example (outlined in Figure 1). These paralogous yeast genes, called MPK1 (for MAP Kinase 1) and MLP1 (for Mpk1-Like Protein), encode proteins whose functions are activated through a complex pathway of protein phosphorylation, known as a MAP kinase cascade. This cascade is activated in response to cell wall stress and is essential to the maintenance of cell wall integrity in fungal species. The Mpk1 protein retains two ancestral functions, one of which is protein kinase activity against targets that include a transcription factor called Rlm1. The second function of Mpk1 is highly atypical– it acts as a co-transcription factor (with another factor called SBF). Both of the Mpk1 functions require its activation by an upstream protein kinase (Mkk1/2), but its co-transcriptional function does not require its catalytic (protein kinase) activity.
Now lets consider Mlp1. Relative to Mpk1 (and all other known MAP kinases, of which there are more than 100), this protein has sustained two critical mutations that interfere with its ability to position ATP in its active site, and hence block its protein kinase activity (Levin, 2005 MMBR 69:262). We refer to Mlp1 as a pseudokinase. Either mutation alone would have been sufficient to block (or at least severely impair) catalytic activity. Despite this catastrophic impairment, Mlp1 has retained its co-transcriptional function, which as noted above, does not depend on catalytic activity. That is to say, its function has become specialized. As an added twist, expression of MLP1 is under the control of the Mpk1-activated transcription factor, Rlm1. This regulation resulted from the MLP1 promoter having acquired an Rlm1-binding site (Jung and Levin, 1999 Mol. Micro. 34:1049), and subjugates the function of Mlp1 to that of Mpk1. The order of these last two steps (mutational loss-of-function and transcriptional control by Rlm1) is not clear, but neither is it important. What is important is that they happened. So, in three incontrovertible stepsgene duplication, followed by two mutations (loss of catalytic activity and acquisition of an Rlm1-binding site), the MLP1 gene and its encoded protein have progressed 75% of the way toward a new level of irreducible complexity. The final step, which may or may not eventually happen, is specialization of Mpk1 through the mutational loss of its co-transcriptional activation of SBF. We have shown that a simple deletion mutation within the MPK1 gene is sufficient to effect this specialization. We are currently preparing this work for publication.
INSERT DIAGRAM: levin_diagram.jpg
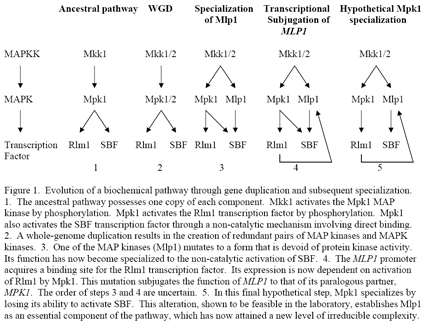
Two aspects of this example are particularly compelling. The first is the inescapable conclusion that MLP1 and MPK1 are paralogous genes that resulted from a duplication event. The second is that only a small number of functionally important alterations were required for the specialization of MLP1 and its functional subjugation to MPK1. From this and other examples cited by a considerable number of critics of the argument from irreducible complexity, it is unmistakably clear that gradual evolution of an irreducibly complex pathway is not only plausible, but it is happening! And its been happening through mechanisms like the one described above for billions of years. Like it or not Dr. Behe, no amount of dissembling on your part is going to change that. Some such pathways are so ancient that the tracks of their evolutionary history have nearly been covered by the sands of time. Nevertheless, the close sequence similarities shared among the proteases of the clotting cascade, the caspases of the apoptotic cascade, and the protein kinases of MAP kinase cascades, to name but a few, are evidence of more than just common descent. They reveal the likely evolutionary origins of the pathways in which these enzymes participate.
On a final note, I must tell you that I was forced to put your book down in disgust at the end of the section entitled What will science do? Your prediction was so laughable as to warrant repeating here.
The theory of intelligent design promises to reinvigorate a field of science grown stale from a lack of viable solutions to dead-end problems. The intellectual competition created by the discovery of design will bring sharper analysis to the professional scientific literature and will require that assertions be backed by hard data. The theory will spark experimental approaches and new hypotheses that would otherwise be untried. A rigorous theory of intelligent design will be a useful tool for the advancement of science in an area that has been moribund for decades.
Oh please, Dr. Behe, would you be so kind as to direct me to the scientific literature on intelligent design that has undoubtedly emerged in the intervening decade since you made this bold prediction? Whats that you say? There is none? Could it be because intelligent design is not a scientific theory? Could it be that it makes no testable predictions? Could it be that it suggests no method for investigation of the presumptive designer? The notion of intelligent design offers us nothing and threatens to obscure the truth. Congratulations, Dr. Behe. You have succeeded in mobilizing a new warrior to the defense of science education from the spread of your particular brand of blinding ignorance.
Sincerely,
David E. Levin, Ph.D.
Professor of Biochemistry
& Molecular Biology
====Nature article pdf Levin sent to Behe and me=====
