NOTE: This post has been updated with an Appendix – VJT.
My post yesterday, Order is not the same thing as complexity: A response to Harry McCall (17 June 2013), seems to have generated a lively discussion, judging from the comments received to date. Over at The Skeptical Zone, Mark Frank has also written a thoughtful response titled, VJ Torley on Order versus Complexity. In today’s post, I’d like to clear up a few misconceptions that are still floating around.
1. In his opening paragraph, Mark Frank writes:
To sum it up – a pattern has order if it can be generated from a few simple principles. It has complexity if it can’t. There are some well known problems with this – one of which being that it is not possible to prove that a given pattern cannot be generated from a few simple principles. However, I don’t dispute the distinction. The curious thing is that Dembski defines specification in terms of a pattern that can generated from a few simple principles. So no pattern can be both complex in VJ’s sense and specified in Dembski’s sense.
Mark Frank appears to be confusing the term, “generated,” with the term. “described.” here. What I wrote in my post yesterday is that a pattern exhibits order if it can be generated by “a short algorithm or set of commands,” and complexity if it can’t be compressed into a shorter pattern by a general law or computer algorithm. Professor William Dembski, in his paper, Specification: The Pattern That Signifies Intelligence, defines specificity in terms of the shortest verbal description of a pattern. On page 16, Dembski defines the function phi_s(T) for a pattern T as “the number of patterns for which S’s semiotic description of them is at least as simple as S’s semiotic description of T” (emphasis mine) before going on to define the specificity sigma as minus the log (to base 2) of the product of phi_s(T) and P(T|H), where P(T|H) is the probability of the pattern T being formed according to “the relevant chance hypothesis that takes into account Darwinian and other material mechanisms” (p. 17). In The Design of Life: Discovering Signs of Intelligence in Biological Systems (The Foundation for Thought and Ethics, Dallas, 2008), Intelligent Design advocates William Dembski and Jonathan Wells define specification as “low DESCRIPTIVE complexity” (p. 320), and on page 311 they explain that descriptive complexity “generalizes Kolmogorov complexity by measuring the size of the minimum description needed to characterize a pattern.”
The definition of order and complexity relates to whether or not a pattern can be generated mathematically by “a short algorithm or set of commands,” rather than whether or not it can be described in a few words. The definition of specificity, on the other hand, relates to whether or not a pattern can be characterized by a brief verbal description. There is nothing that prevents a pattern from being difficult to generate algorithmically, but easy to describe verbally. Hence it is quite possible for a pattern to be both complex and specified.
NOTE: I have substantially revised my response to Mark Frank, in the Appendix below.
2. Dr. Elizabeth Liddle, in a comment on Mark Frank’s post, writes that “by Dembski’s definition a chladni pattern would be both specified and complex. However, it would not have CSI because it is highly probable given a relevant chance (i.e. non-design) hypothesis.” The second part of her comment is correct; the first part is incorrect. Precisely because a Chladni pattern is “highly probable given a relevant chance (i.e. non-design) hypothesis,” it is not complex. In The Design of Life: Discovering Signs of Intelligence in Biological Systems (The Foundation for Thought and Ethics, Dallas, 2008), William Dembski and Jonathan Wells define complexity as “The degree of difficulty to solve a problem or achieve a result,” before going on to add: “The most common forms of complexity are probabilistic (as in the probability of obtaining some outcome) or computational (as in the memory or computing time required for an algorithm to solve a problem)” (pp. 310-311). If a Chladni pattern is easy to generate as a result of laws then it exhibits order rather than complexity.
3. In another comment, Dr. Liddle writes: “V J Torley seems to be forgetting that fractal patterns are non-repeating, even though they can be simply described.” I would beg to differ. Here’s what Wikipedia has to say in its article on fractals (I’ve omitted references):
Fractals are typically self-similar patterns, where self-similar means they are “the same from near as from far”. Fractals may be exactly the same at every scale, or, as illustrated in Figure 1, they may be nearly the same at different scales. The definition of fractal goes beyond self-similarity per se to exclude trivial self-similarity and include the idea of a detailed pattern repeating itself.
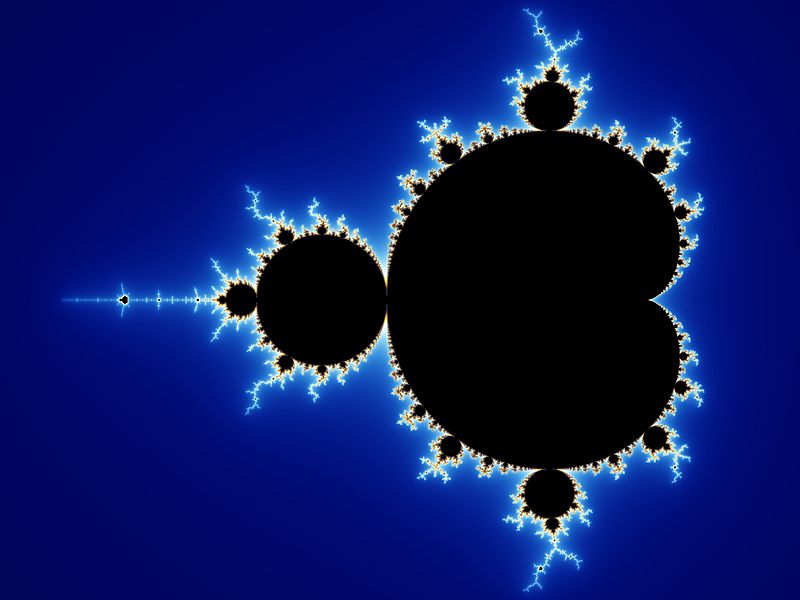 |
The caption accompanying the figure referred to above reads as follows: “The Mandelbrot set illustrates self-similarity. As you zoom in on the image at finer and finer scales, the same pattern re-appears so that it is virtually impossible to know at which level you are looking.”
That sounds pretty repetitive to me. More to the point, fractals are mathematically easy to generate. Here’s what Wikipedia says about the Mandelbrot set, for instance:
More precisely, the Mandelbrot set is the set of values of c in the complex plane for which the orbit of 0 under iteration of the Complex quadratic polynomial zn+1 = zn2 + c remains bounded. That is, a complex number c is part of the Mandelbrot set if, when starting with z0 = 0 and applying the iteration repeatedly, the absolute value of zn remains bounded however large n gets.
NOTE: I have revised some of my comments on Mandelbrot sets and fractals. See the Appendix below.
4. In another comment on the same post, Professor Joe Felsenstein objects that Dembski’s definition of specified complexity has a paradoxical consequence: “It implies that we are to regard a life form as uncomplex, and therefore having specified complexity [?] if it is easy to describe,” which means that “a hummingbird, on that view, has not nearly as much specification as a perfect steel sphere,” even though the hummingbird “can do all sorts of amazing things, including reproduce, which the steel sphere never will.” He then suggests defining specification on a scale of fitness.
In my post yesterday, I pointed out that the term “specified complexity” is fairly non-controversial when applied to life: as chemist Leslie Orgel remarked in 1973, “living organisms are distinguished by their specified complexity.” Orgel added that crystals are well-specified, but simple rather than complex. If specificty were defined in terms of fitness, as Professor Felsenstein suggests, then we could no longer say that a non-reproducing crystal was specified.
However, Professor Felsenstein’s example of the steel sphere is an interesting one, because it illustrates that the probability of a sphere’s originating by natural processes may indeed be extremely low, especially if it is also made of an exotic material. (In this respect, it is rather like the lunar monolith in the movie 2001.) Felsenstein’s point is that a living organism would be a worthier creation of an intelligent agent than such a sphere, as it has a much richer repertoire of capabilities.
Closely related to this point is the fact that living things exhibit a nested hierarchy of organization, as well as dedicated functionality: intrinsically adapted parts whose entire repertoire of functionality is “dedicated” to supporting the functionality of the whole unit which they comprise. Indeed, it is precisely this kind of organization and dedicated functionality which allows living things to reproduce in the first place.
At the bottom level, the full biochemical specifications required for putting together a living thing such as a hummingbird are very long indeed. It is only when we get to higher organizational levels that we can apply holistic language and shorten our description, by characterizing the hummingbird in terms of its bodily functions rather than its parts, and by describing those functions in terms of how they benefit the whole organism.
I would therefore agree that an entity exhibiting this combination of traits (bottom-level exhaustive detail and higher-level holistic functionality, which makes the entity easy to characterize in a few words) is a much more typical product of intelligent agency than a steel sphere, notwithstanding the latter’s descriptive simplicity.
In short: specified complexity gets us to Intelligent Design, but some designs are a lot more intelligent than others. Whoever made hummingbirds must have been a lot smarter than we are; we have enough difficulties putting together a single protein.
5. In a comment on my post, Alan Fox objects that “We simply don’t know how rare novel functional proteins are.” Here I should refer him to the remarks made by Dr. Branko Kozulic in his 2011 paper, Proteins and Genes, Singletons and Species. I shall quote a brief extract:
In general, there are two aspects of biological function of every protein, and both depend on correct 3D structure. Each protein specifically recognizes its cellular or extracellular counterpart: for example an enzyme its substrate, hormone its receptor, lectin sugar, repressor DNA, etc. In addition, proteins interact continuously or transiently with other proteins, forming an interactive network. This second aspect is no less important, as illustrated in many studies of protein-protein interactions [59, 60]. Exquisite structural requirements must often be fulfilled for proper functioning of a protein. For example, in enzymes spatial misplacement of catalytic residues by even a few tenths of an angstrom can mean the difference between full activity and none at all [54]. And in the words of Francis Crick, “To produce this miracle of molecular construction all the cell need do is to string together the amino acids (which make up the polypeptide chain) in the correct order”….
Let us assess the highest probability for finding this correct order by random trials and call it, to stay in line with Crick’s term, a “macromolecular miracle”. The experimental data of Keefe and Szostak indicate – if one disregards the above described reservations – that one from a set of 10^11 randomly assembled polypeptides can be functional in vitro, whereas the data of Silverman et al. [57] show that of the 10^10 in vitro functional proteins just one may function properly in vivo. The combination of these two figures then defines a “macromolecular miracle” as a probability of one against 10^21. For simplicity, let us round this figure to one against 10^20…
It is important to recognize that the one in 10^20 represents the upper limit, and as such this figure is in agreement with all previous lower probability estimates. Moreover, there are two components that contribute to this figure: first, there is a component related to the particular activity of a protein – for example enzymatic activity that can be assayed in vitro or in vivo – and second, there is a component related to proper functioning of that protein in the cellular context: in a biochemical pathway, cycle or complex. (pp. 7-8)
In short: the specificity of proteins is not in doubt, and their usefulness for Intelligent Design arguments is therefore obvious.
I sincerely hope that the foregoing remarks will remove some common misunderstandings and stimulate further discussion.
APPENDIX
Let me begin with a confession: I had a nagging doubt when I put up this post a couple of days ago. What bothered me was that (a) some of the definitions of key terms were a little sloppily worded; and (b) some of these definitions seemed to conflate mathematics with physics.
Maybe I should pay more attention to my feelings.
A comment by Professor Jeffrey Shallit over at The Skeptical Zone also convinced me that I needed to re-think my response to Mark Frank on the proper definition of specificity. Professor Shallit’s remarks on Kolmogorov complexity also made me realize that I needed to be a lot more careful about defining the term “generate,” which may denote either a causal process governed by physical laws, or the execution of an algorithm by performing a sequence of mathematical operations.
What I wrote in my original post, Order is not the same thing as complexity: A response to Harry McCall (17 June 2013), is that a pattern exhibits order if it can be generated by “a short algorithm or set of commands,” and complexity if it can’t be compressed into a shorter pattern by a general law or computer algorithm.
I’d now like to explain why I now find those definitions unsatisfactory, and what I would propose in their stead.
Problems with the definition of order
I’d like to start by going back to the original sources. In Signature in the Cell l (Harper One, 2009, p. 106), Dr. Stephen Meyer writes:
Complex sequences exhibit an irregular, nonrepeating arrangement that defies expression by a general law or computer algorithm (an algorithm is a set of expressions for accomplishing a specific task or mathematical operation). The opposite of a highly complex sequence is a highly ordered sequence like ABCABCABCABC, in which the characters or constituents repeat over and over due to some underlying rule, algorithm or general law. (p. 106)
[H]igh probability repeating sequences like ABCABCABCABCABCABC have very little information (either carrying capacity or content)… Such sequences aren’t complex either. Why? A short algorithm or set of commands could easily generate a long sequence of repeating ABC’s, making the sequence compressible. (p. 107)
(Emphases mine – VJT.)
There are two problems with this definition. First, it mistakenly conflates physics with mathematics, when it declares that a complex sequence can be generated by “a general law or computer algorithm.” I presume that by “general law,” Dr. Meyer means to refer to some law of Nature, since on page 107, he lists certain kinds of organic molecules as examples of complexity. The problem here is that a sequence may be easy to generate by a computer algorithm, but difficult to generate by the laws of physics (or vice versa). In that case, it may be complex according to physical criteria but not according to mathematical criteria (or the reverse), generating a contradiction.
Second, the definition conflates: (a) the repetitiveness of a sequence, with (b) the ability of a short algorithm to generate that sequence, and (c) the Shannon compressibility of that sequence. The problem here is that there are non-repetitive sequences which can be generated by a short algorithm. Some of these non-repeating sequences are also Shannon-incompressible. Do these sequences exhibit order or complexity?
Third, the definition conflicts with what Professor Dembski has written on the subject of order and complexity. In The Design of Life: Discovering Signs of Intelligence in Biological Systems (The Foundation for Thought and Ethics, Dallas, 2008), Professor William Dembski and Dr. Jonathan Wells provide three definitions for order, the first of which reads as follows:
(1) Simple or repetitive patterns, as in crystals, that are the result of laws and cannot reasonably be used to draw a design inference. (p. 317; italics mine – VJT).
The reader will notice that the definition refers only to law-governed physical processes.
Dembski’s 2005 paper, Specification: The Pattern that Signifies Intelligence, also refers to the Champernowne sequence as exhibiting a “combination of pattern simplicity (i.e., easy description of pattern) and event-complexity (i.e., difficulty of reproducing the corresponding event by chance)” (pp. 15-16). According to Dembski, the Champernowne sequence can be “constructed simply by writing binary numbers in ascending lexicographic order, starting with the one-digit binary
numbers (i.e., 0 and 1), proceeding to the two-digit binary numbers (i.e., 00, 01, 10, and 11),” and so on indefinitely, which means that it can be generated by a short algorithm. At the same time, Dembski describes at as having “event-complexity (i.e., difficulty of reproducing the corresponding event by chance).” In other words, it is not an example of what he would define as order. And yet, because it can be generated by “a short algorithm,” it would arguably qualify as an example of order under Dr. Meyer’s criteria (see above).
Problems with the definition of specificity
Dr. Meyer’s definition of specificity is also at odds with Dembski’s. On page 96 of Signature in the Cell, Dr. Meyer defines specificity in exclusively functional terms:
By specificity, biologists mean that a molecule has some features that have to be what they are, within fine tolerances, for the molecule to perform an important function within the cell.
Likewise, on page 107, Meyer speaks of a sequence of digits as “specifically arranged to form a function.”
By contrast, in The Design of Life: Discovering Signs of Intelligence in Biological Systems (The Foundation for Thought and Ethics, Dallas, 2008), Professor William Dembski and Dr. Jonathan Wells define specification as “low DESCRIPTIVE complexity” (p. 320), and on page 311 they explain that descriptive complexity “generalizes Kolmogorov complexity by measuring the size of the minimum description needed to characterize a pattern.” Although Dembski certainly regards functional specificity as one form of specificity, since he elsewhere refers to the bacterial flagellum – a “bidirectional rotary motor-driven propeller” – as exhibiting specificity, he does not regard it as the only kind of specificity.
In short: I believe there is a need for greater rigor and consistency when defining these key terms. Let me add that I don’t wish to criticize any of the authors I’ve mentioned above; I’ve been guilty of terminological imprecision at times, myself.
My suggestions for more rigorous definitions of the terms “order” and “specification”
So here are my suggestions. In Specification: The Pattern that Signifies Intelligence, Professor Dembski defines a specification in terms of a “combination of pattern simplicity (i.e., easy description of pattern) and event-complexity (i.e., difficulty of reproducing the corresponding event by chance),” and in The Design of Life: Discovering Signs of Intelligence in Biological Systems (The Foundation for Thought and Ethics, Dallas, 2008), Dembski and Wells define complex specified information as being equivalent to specified complexity (p. 311), which they define as follows:
An event or object exhibits specified complexity provided that (1) the pattern to which it conforms is a highly improbable event (i.e. has high PROBABILISTIC COMPLEXITY) and (2) the pattern itself is easily described (i.e. has low DESCRIPTIVE COMPLEXITY). (2008, p. 320)
What I’d like to propose is that the term order should be used in opposition to high probabilistic complexity. In other words, a pattern is ordered if and only if its emergence as a result of law-governed physical processes is not a highly improbable event. More succinctly: a pattern is ordered is it is reasonably likely to occur, in our universe, and complex if its physical realization in our universe is a very unlikely event.
Thus I was correct when I wrote above:
If a Chladni pattern is easy to generate as a result of laws then it exhibits order rather than complexity.
However, I was wrong to argue that a repeating pattern is necessarily a sign of order. In a salt crystal it certainly is; but in the sequence of rolls of a die, a repeating pattern (e.g. 123456123456…) is a very improbable pattern, and hence it would be probabilistically complex. (It is, of course, also a specification.)
Fractals, revisited
The same line of argument holds true for fractals: when assessing whether they exhibit order or (probabilistic) complexity, the question is not whether they repeat themselves or are easily generated by mathematical algorithms, but whether or not they can be generated by law-governed physical processes. I’ve seen conflicting claims on this score (see here and here and here): some say there are fractals in Nature, while other say that some objects in Nature have fractal features, and still others, that the patterns that produce fractals occur in Nature even if fractals themselves do not. I’ll leave that one to the experts to sort out.
The term specification should be used to refer to any pattern of low descriptive complexity, whether functional or not. (I say this because some non-functional patterns, such as the lunar monolith in 2001, and of course fractals, are clearly specified.)
Low Kolmogorov complexity is, I would argue, a special case of specification. Dembski and Wells agree: on page 311 of The Design of Life: Discovering Signs of Intelligence in Biological Systems (The Foundation for Thought and Ethics, Dallas, 2008), they explain that descriptive complexity “generalizes Kolmogorov complexity by measuring the size of the minimum description needed to characterize a pattern” (italics mine).
Kolmogorov complexity as a special case of descriptive complexity
Which brings me to Professor Shallit’s remarks in a post over at The Skeptical Zone, in response to my earlier (misguided) attempt to draw a distinction between the mathematical generation of a pattern and the verbal description of that pattern:
In the Kolmogorov setting, “concisely described” and “concisely generated” are synonymous. That is because a “description” in the Kolmogorov sense is the same thing as a “generation”; descriptions of an object x in Kolmogorov are Turing machines T together with inputs i such that T on input i produces x. The size of the particular description is the size of T plus the size of i, and the Kolmogorov complexity is the minimum over all such descriptions.
I accept Professor Shallit’s correction on this point. What I would insist, however, is that the term “descriptive complexity,” as used by the Intelligent design movement, cannot be simply equated with Kolmogorov complexity. Rather, I would argue that low Kolmogorov complexity is a special case of low descriptive complexity. My reason for adopting this view is that the determination of a object’s Kolmogorov complexity requires a Turing machine (a hypothetical device that manipulates symbols on a strip of tape according to a table of rules), which is an inappropriate (not to mention inefficient) means of determining whether an object possesses functionality of a particular kind – e.g. is this object a cutting implement? What I’m suggesting, in other words, is that at least some functional terms in our language are epistemically basic, and that our recognition of whether an object possesses these functions is partly intuitive. Using a table of rules to determine whether or not an object possesses a function (say, cutting) is, in my opinion, likely to produce misleading results.
My response to Mark Frank, revisited
I’d now like to return to my response to Mark Frank above, in which I wrote:
The definition of order and complexity relates to whether or not a pattern can be generated mathematically by “a short algorithm or set of commands,” rather than whether or not it can be described in a few words. The definition of specificity, on the other hand, relates to whether or not a pattern can be characterized by a brief verbal description. There is nothing that prevents a pattern from being difficult to generate algorithmically, but easy to describe verbally. Hence it is quite possible for a pattern to be both complex and specified.
This, I would now say, is incorrect as it stands. The reason why it is quite possible for an object to be both complex and specified is that the term “complex” refers to the (very low) likelihood of its originating as a result of physical laws (not mathematical algorithms), whereas the term “specified” refers to whether it can be described briefly – whether it be according to some algorithm or in functional terms.
Implications for Intelligent Design
I have argued above that we can legitimately infer an Intelligent Designer for any system which is capable of being verbally described in just a few words, and whose likelihood of originating as a result of natural laws is sufficiently close to zero. This design inference is especially obvious in systems which exhibit biological functionality. Although we can make design inferences for non-biological systems (e.g. moon monoliths, if we found them), the most powerful inferences are undoubtedly drawn from the world of living things, with their rich functionality.
In an especially perspicuous post on this thread, G. Puccio argued for the same conclusion:
The simple truth is, IMO, that any kind of specification, however defined, will do, provided that we can show that that specification defines a specific subset of the search space that is too small to be found by a random search, and which cannot reasonably be found by some natural algorithm….
In the end, I will say it again: the important point is not how you specify, but that your specification identifies:
a)an utterly unlikely subset as a pre-specification
or
b) an utterly unlikely subset which is objectively defined without any arbitrary contingency, like in the case of pi.
In the second case, specification needs not be a pre-specification.
Functional specification is a perfect example of the second case.
Provided that the function can be objectively defined and measured, the only important point is how complex it is: IOWs, how small is the subset of sequences that provide the function as defined, in the search space.
That simple concept is the foundation for the definition of dFSCI, or any equivalent metrics.
It is simple, it is true, it works.
P{T|H) and elephants: Dr. Liddle objects
But how do we calculate probabilistic complexities? Dr. Elizabeth Liddle writes:
P(T|H) is fine to compute if you have a clearly defined non-design hypothesis for which you can compute a probability distribution.
But nobody, to my knowledge, has yet as suggested how you would compute it for a biological organism, or even for a protein.
In a similar vein, Alan Fox comments:
We have, as yet, no way to predict functionality in unknown proteins. Without knowing what you don’t know, you can’t calculate rarity.
In a recent post entitled, The Edge of Evolution, I cited a 2011 paper by Dr. Branko Kozulic, titled, Proteins and Genes, Singletons and Species, in which he argued (generously, in his view) that at most, 1 in 10^21 randomly assembled polypeptides would be capable of functioning as a viable protein in vivo, that each species possessed hundreds of isolated proteins called “singletons” which had no close biochemical relatives, and that the likelihood of these proteins originating by unguided mechanisms in even one species was astronomically low, making proteins at once highly complex (probabilistically speaking) and highly specified (by virtue of their function) – and hence as sure a sign as we could possibly expect of an Intelligent Designer at work in the natural world:
In general, there are two aspects of biological function of every protein, and both depend on correct 3D structure. Each protein specifically recognizes its cellular or extracellular counterpart: for example an enzyme its substrate, hormone its receptor, lectin sugar, repressor DNA, etc. In addition, proteins interact continuously or transiently with other proteins, forming an interactive network. This second aspect is no less important, as illustrated in many studies of protein-protein interactions [59, 60]. Exquisite structural requirements must often be fulfilled for proper functioning of a protein. For example, in enzymes spatial misplacement of catalytic residues by even a few tenths of an angstrom can mean the difference between full activity and none at all [54]. And in the words of Francis Crick, “To produce this miracle of molecular construction all the cell need do is to string together the amino acids (which make up the polypeptide chain) in the correct order” [61, italics in original]. (pp. 7-8)
Let us assess the highest probability for finding this correct order by random trials and call it, to stay in line with Crick’s term, a “macromolecular miracle”. The experimental data of Keefe and Szostak indicate – if one disregards the above described reservations – that one from a set of 10^11 randomly assembled polypeptides can be functional in vitro, whereas the data of Silverman et al. [57] show that of the 10^10 in vitro functional proteins just one may function properly in vivo. The combination of these two figures then defines a “macromolecular miracle” as a probability of one against 10^21. For simplicity, let us round this figure to one against 10^20. (p. 8)
To put the 10^20 figure in the context of observable objects, about 10^20 squares each measuring 1 mm^2 would cover the whole surface of planet Earth (5.1 x 10^14 m^2). Searching through such squares to find a single one with the correct number, at a rate of 1000 per second, would take 10^17 seconds, or 3.2 billion years. Yet, based on the above discussed experimental data, one in 10^20 is the highest probability that a blind search has for finding among random sequences an in vivo functional protein. (p. 9)
The frequency of functional proteins among random sequences is at most one in 10^20 (see above). The proteins of unrelated sequences are as different as the proteins of random sequences [22, 81, 82] – and singletons per definition are exactly such unrelated proteins. (p. 11)
A recent study, based on 573 sequenced bacterial genomes, has concluded that the entire pool of bacterial genes – the bacterial pan-genome – looks as though of infinite size, because every additional bacterial genome sequenced has added over 200 new singletons [111]. In agreement with this conclusion are the results of the Global Ocean Sampling project reported by Yooseph et al., who found a linear increase in the number of singletons with the number of new protein sequences, even when the number of the new sequences ran into millions [112]. The trend towards higher numbers of singletons per genome seems to coincide with a higher proportion of the eukaryotic genomes sequenced. In other words, eukaryotes generally contain a larger number of singletons than eubacteria and archaea. (p. 16)
Based on the data from 120 sequenced genomes, in 2004 Grant et al. reported on the presence of 112,000 singletons within 600,000 sequences [96]. This corresponds to 933 singletons per genome…
[E]ach species possesses hundreds, or even thousands, of unique genes – the genes that are not shared with any other species. (p. 17)Experimental data reviewed here suggest that at most one functional protein can be found among 10^20 proteins of random sequences. Hence every discovery of a novel functional protein (singleton) represents a testimony for successful overcoming of the probability barrier of one against at least 10^20, the probability defined here as a “macromolecular miracle”. More than one million of such “macromolecular miracles” are present in the genomes of about two thousand species sequenced thus far. Assuming that this correlation will hold with the rest of about 10 million different species that live on Earth [157], the total number of “macromolecular miracles” in all genomes could reach 10 billion. These 10^10 unique proteins would still represent a tiny fraction of the 10^470 possible proteins of the median eukaryotic size. (p. 21)
If just 200 unique proteins are present in each species, the probability of their simultaneous appearance is one against at least 10^4,000. [The] Probabilistic resources of our universe are much, much smaller; they allow for a maximum of 10^149 events [158] and thus could account for a one-time simultaneous appearance of at most 7 unique proteins. The alternative, a sequential appearance of singletons, would require that the descendants of one family live through hundreds of “macromolecular miracles” to become a new species – again a scenario of exceedingly low probability. Therefore, now one can say that each species is a result of a Biological Big Bang; to reserve that term just for the first living organism [21] is not justified anymore. (p. 21)
“But what if the search for a functional protein is not blind?” ask my critics. “What if there’s an underlying bias towards the emergence of functionality in Nature?” “Fine,” I would respond. “Let’s see your evidence.”
Alan Miller rose to the challenge. In a recent post entitled, Protein Space and Hoyle’s Fallacy – a response to vjtorley, cited a paper by Michael A. Fisher, Kara L. McKinley, Luke H. Bradley, Sara R. Viola and Michael H. Hecht, titled, De Novo Designed Proteins from a Library of Artificial Sequences Function in Escherichia Coli and Enable Cell Growth (PLoS ONE 6(1): e15364. doi:10.1371/journal.pone.0015364, January 4, 2011), in support of his claim that proteins were a lot easier for Nature to build on the primordial Earth than Intelligent Design proponents imagine, and he accused them of resurrecting Hoyle’s fallacy.
In a very thoughtful comment over on my post CSI Revisited, G. Puccio responded to the key claims made in the paper, and to what he perceived as Alan Miller’s misuse of the paper (bolding below is mine):
First of all, I will just quote a few phrases from the paper, just to give the general scenario of the problems:
a) “We designed and constructed a collection of artificial genes encoding approximately 1.5×106 novel amino acid sequences. Because folding into a stable 3-dimensional structure is a prerequisite for most biological functions, we did not construct this collection of proteins from random sequences. Instead, we used the binary code strategy for protein design, shown previously to facilitate the production of large combinatorial libraries of folded proteins.”
b) “Cells relying on the de novo proteins grow significantly slower than those expressing the natural protein.”
c) “We also purified several of the de novo proteins. (To avoid contamination by the natural enzyme, purifications were from strains deleted for the natural gene.) We tested these purified proteins for the enzymatic activities deleted in the respective autotrophs, but were unable to detect activity that was reproducibly above the controls.”
And now, my comments:
a) This is the main fault of the paper, if it is interpreted (as Miller does) as evidence that functional proteins can evolve from random sequences. The very first step of the paper is intelligent design: indeed, top down protein engineering based on our hardly gained knowledge about the biochemical properties of proteins.
b) The second problem is that the paper is based on function rescue, not on the appearance of a mew function. Experiments based on function rescue have serious methodological problems, if used as models of neo darwinian evolution. The problem here is specially big, because we know nothing of how the “evolved” proteins work to allow the minimal rescue of function in the complex system of E. Coli (see next point).
c) The third problem is that the few rescuing sequences have no detected biochemical activity in vitro. IOWs, we don’t know what they do, and how they act at biochemical level. IOWs, with no known “local function” for the sequences, we have no idea of the functional complexity of the “local function” that in some unknown way is linked to the functional rescue. The authors are well aware of that, and indeed spend a lot of time discussing some arguments and experiments to exclude some possible interpretation of indirect rescue, or at least those that they have conceived.
The fact remains that the hypothesis that the de novo sequences have the same functional activity as the knocked out genes, even if minimal, remain unproved, because no biochemical activity of that kind could be shown in vitro for them.
These are the main points that must be considered. In brief, the paper does not prove, in any way, what Miller thinks it proves.
And that was Miller’s best paper!
In the meantime, can you forgive us in the Intelligent Design community for being just a little skeptical of claims that “no intelligence was required” to account for the origin of proteins, of the first living cell (which would have probably required hundreds of proteins), of complex organisms in the early Cambrian period, and even of the appearance of a new species, in view of what has been learned about the prevalence of singleton proteins and genes in living organisms?