In eukaryotic organisms and especially humans, large amounts of the DNA are transcribed into RNAs that never end up getting translated into proteins. This has led some to argue human DNA is mostly junk. I speculate otherwise. The supposed junk DNA that transcribes to supposed junk RNA is not junk at all.
I recently discovered that RNA is an excellent chemical basis for molecular level sensing, logic, computation and communication. Because of these facts I speculate RNAs are important in navigation, organization, computation and processing of information in the eukaryotic cell. I contrast my view with that Darwinist Steve Matheson who debated Stephen Meyer regarding claims in Meyer’s book, Signature in the Cell.
Matheson had a dismissive view of DNAs that made RNAs that didn’t leave the nucleus:
they’re snipped out of the transcripts and discarded before the darn things even leave the nucleus.
Did it ever occur to Matheson these RNAs actually did something in the nucleus? DUH! See below for possible hints as to why some RNAs need to stay inside the nucleus…
Consider a car GPS system that gives a simple instruction of “turn left” or “turn right”. It must do a few tasks in order to issue these instructions. It needs to determine the present position and what road the driver is on, it needs to compute a route and then compute what instructions to give. Even more complex is a computerized Air Traffic Control decision support system that does enormous amounts of computation to determine when and how each aircraft must make a maneuver.
Like managing air traffic, similar challenges exist in controlling the traffic of 1 billion physical proteins in a cell. Something as simple as an instruction to express or not express a protein, to specify a location for the protein, etc. might entail a fair amount of computation to make what superficially seems trivial instructions for each individual protein. And there must also be computation in order to manage communications with other other cells so that an organism like a human consisting of 200 billion trillion proteins can function in a coordinated fashion.
With the need of fair amounts of sensing, logic, communication and computation required to develop a human being, and with the yet unaccounted functions of non-coding RNAs, is it possible these RNAs meet some of these needs? So I asked the question, how ideal are RNAs for making such nano-sensing, logic, computation and communication?
Answer: very good! See: Computing with RNA.
Scientists in California have created molecular computers that are able to self-assemble out of strips of RNA within living cells. Eventually, such computers could be programmed to manipulate biological functions within the cell, executing different tasks under different conditions. One application could be smart drug delivery systems, says Christina Smolke, who carried out the research with Maung Nyan Win and whose results are published in the latest issue of Science.
The use of biomolecules to perform computations was first demonstrated by the University of Southern California’s Leonard Adleman in 1994, and the approach was later developed by Ehud Shapiro of the Weizmann Institute of Science, in Rehovot, Israel. But according to Shapiro, “What this new work shows for the first time is the ability to detect the presence or absence of molecules within the cell.”
That opens up the possibility of computing devices that can respond to specific conditions within the cell, he says. For example, it may be possible to develop drug delivery systems that target cancer cells from within by sensing genes used to regulate cell growth and death. “You can program it to release the drug when the conditions are just right, at the right time and in the right place,” Shapiro says.
Smolke and Win’s biocomputers are built from three main components–sensors, actuators, and transmitters–all of which are made up of RNA. The input sensors are made from aptamers, RNA molecules that behave a bit like antibodies, binding tightly to specific targets. Similarly, the output components, or actuators, are made of ribozymes, complex RNA molecules that have catalytic properties similar to those of enzymes. These two components are joined by yet another RNA molecule that serves as a transmitter, which is activated when a sensor molecule recognizes an input chemical and, in turn, triggers an actuator molecule.
It seems to me, junk DNA advocates are underappreciating the computational and information processing requirements to build something as complex as a human beings. Building a human being is more than just making proteins and stirring them together! If all it took to make life was having lots of proteins and stirring them together, then several frogs mixed in a blender with broccoli could make a novel life form.
Since RNAs are ideal components for nano information processing machines, they may be indeed the materials of such machines in cells and not merely junk as Steve Matheson insinuated, but a centerpiece of computation and control.
NOTES
Consider this description from the Nobel Prize website of Ribosome Assembly:
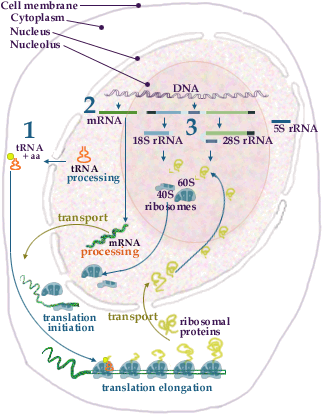
Ribosome Assembly I
Ribosome assembly takes place in the nucleolus. The assembly of the 40S subunit (exemplified in the illustration) starts as the giant primary transcript is being produced. During this period initial folding and snoRNA-dependent cleavage of the primary transcript take place. Other snoRNAs direct modification of specific nucleotides in the rRNA. Primary rRNA binding ribosomal proteins associate to the rRNA and the folding of the rRNA proceeds. The next set of ribosomal proteins, i.e. proteins which do not bind directly to rRNA, join the rRNA-protein complex thereby forming the 40S ribosomal subunit. For production of the 60S subunit the assembly process also has to be coordinated with the production of 5S rRNA. The whole ribosome synthesis and assembly process is an intricate interplay between the nucleolus, the nucleus and the cytoplasm.
Ribosome Assembly
1. tRNAs are transcribed in the nucleoplasm by RNA polymerase III. The primary transcript is processed to the mature tRNA and transported to the cytoplasm. In the cytoplasm the tRNAs are coupled to their cognate amino acids.
2. Pre-mRNAs, containing the sequences coding for the ribosomal proteins, are transcribed by RNA polymerase II in the nucleoplasm. The pre-mRNAs are processed (see splicing) and transported (see transport) as mRNP-particles to the cytoplasm where they are picked up by the ribosomes
initiation and translated elongation into ribosomal proteins. The mature ribosomal proteins are then transported (see transport) to the nucleolus.3. The ribosomal RNAs (except 5S rRNA) are transcribed in the nucleolus as one giant precursor RNA by RNA polymerase I. The precursor is processed (see rRNA processing) to 18S,5.8S and 28S rRNA. 5S rRNA is transcribed in the nucleoplasm by RNA polymerase III and transported to the nucleolus. The rRNAs are folded and associate with ribosomal proteins to form the 40S and 60S ribosomal subunits. The subunits are then transported from the nucleolus to the cytoplasm.
An example of the Rate of Ribosome Synthesis:
*HeLa cells (a type of human tumour cells) divide each 24 hours.
* Each cell contains around 10 million ribosomes, i.e. 7000 ribosomes are produced in the nucleolus each minute.
*Each ribosome contains around 80 proteins, i.e. more than 0.5 million ribosomal proteins are synthesised in the cytoplasm per minute.
*The nuclear membrane contains approximately 5000 pores. Thus, more than 100 ribosomal proteins are imported from the cytoplasm to the nucleus per pore and minute. At the same time 3 ribosomal subunits are exported from the nucleus to the cytoplasm per pore and minute.
So we see that there is manufacturing inside the cell nucleus! It’s not too much of a stretch to think many of the untranslated RNAs that never leave the nucleus might serve an important part in influencing the manufacture of ribosomes inside the nucleus.
2. The “Vodka!” designation is for highly speculative ideas that are likely wrong but may have an important grain of truth.
3. Photo Credits: Nobel Prize website
4. One air traffic control decision support tool is URET.