[cross posted at CEU IDCS, Neutral theory and non-Darwinian evolution for newbies, Part 2]
Part 1 laid out the claim that most nucleotides in populations cannot as a matter of principle be under strong selection, but must be neutral. MOST certainly does not mean ALL. Clearly some deleterious traits if they appear would be lethal, and conversely in certain contexts like antibiotic and pesticide resistance, some traits can be strongly selected for, but these cases do not speak for most of the rest of the molecules in various species. As one scientist said:
Most molecular evolution is neutral. Done.
PZ Myers
Part 2 will focus on how neutral or nearly neutral traits in small finite populations get “fixed” where the word “fixed” in population genetics is used to describe a trait that has practically become part of every individual and is virtually a permanent feature of the entire species.
The Darwinian view in the 1950s was that most of the genome and features of populations were the result of selection. But smart geneticists like Kimura (and unwittingly Haldane) realized that this could not be possible even in principle. This was explained in Part 1.
The present state of affairs is that both evolutionists and creationists with background in population genetics agree that IF we evolved, most of the traits became fixed without the influence of selection. Thus most evolution, if it is unguided and not front loaded, would essentially be random.
These considerations contradict Dawkins’ claim:
You often find people who say, well, evolution is a theory of chance, in the absence of a designer. If it really were a theory of chance, of course they would be right to dismiss it as nonsense. No chance process could give rise to the prodigy of organized complexity that is the living world. But it’s not random chance. Natural selection is the exact opposite of a chance process.
The reason Dawkins is wrong is that if there was evolution and it was unguided, because most molecular evolution is neutral, Darwinian selection has almost no role, and chance would be the only mechanism left. Thus I would actually agree with Dawkins on this:
You often find people who say, well, evolution is a theory of chance, in the absence of a designer. If it really were a theory of chance, of course they would be right to dismiss it as nonsense.
Absence of selection shows unguided evolution is mostly a theory of chance. The proportion of molecular “traits” fixed by selection versus those fixed by random chance were investigated by members of the Mendel’s Accountant team. Their finding are in agreement with claims which are accepted by many scientists in relevant fields but which are not broadcast to the general public, certainly not by Dawkins.
In Using Numerical Simulation to Better Understand Fixation Rates, Rupe and Sanford show in one simulation that even with strong selection against deleterious traits, the ratio of deleterious traits that got fixed by chance versus the beneficial traits that got fixed by Darwinian selection was 6,775 to 10. This figure absolutely refuted Darwin’s fantasy of never ending evolutionary progress. This showed over time that even though some beneficial traits got fixed into the population (like maybe the color of a peppered moth), for every beneficial trait that got locked into the population, 676 bad traits got locked in as well!
So how can this possibly be? Isn’t Darwinian selection supposed to sort out the good from the bad? Well yes if that was the way nature really worked. But the only place Darwinian selection works is not in nature but in genetic algorithms that have no resemblance to biological reality — algorithms such as those implemented in Dawkins Weasel, Avida, Tierra, Ev, Steiner, Geometric and Cordova’s Remarkable Algorithm. Nature doesn’t work this way. Why?
First of all, in sexually reproducing populations, an otherwise advantageous trait can be lost just to random chance. We often observe that some children don’t inherit some of their parents traits. This is because of recombination during meiosis. For example, if Dad has one point mutational “trait” in a particular nucleotide on one chromosome, and Mom does not have that same “trait”, there is only a 25% chance that this otherwise advantageous trait will appear in any given child. And then if a child has that trait, there is no guarantee he might not die or fail to reproduce from accidents or other chance events. Thus chance has a huge role in what persists or gets lost in a population even if a trait might otherwise have a selective advantage. But since most molecular “traits” are neutral, what traits end up persisting in a population are mostly decided by chance.
On the website inbreeding and neutral evolution we see how quickly fixation happens in a small inbreeding population. The ideas for small inbreeding populations can be somewhat scaled up to larger populations provided that the individuals stir well with each other rather than stay isolated for too long. I focus on small populations to help the reader conceptualize fixation under neutral conditions:
Below is an example of drift. Imagine a rare species kept in a zoo with a population of only six diploid individuals. There are a total of 12 alleles (numbered 1-12 in generation 0). All alleles are assumed equally fit, so that evolution is neutral. The alleles may also be genetically distinguishable, or “different in state” (represented by colours).
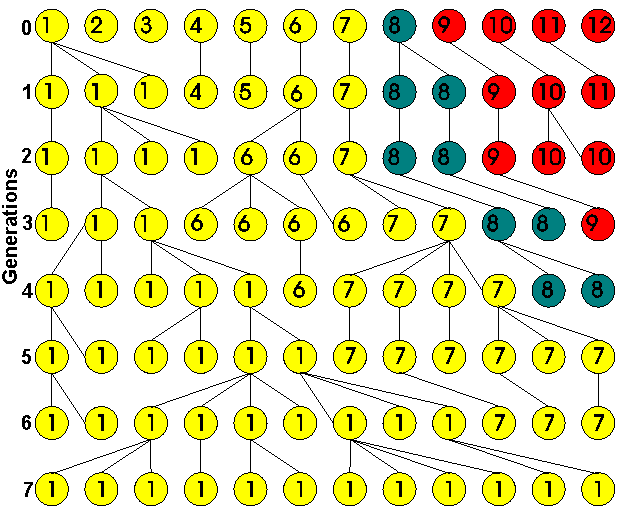
By generation 7, every allele has become identical by a chance processes alone.
This kind of evolution is not predictable; it is random or stochastic.
At the end of several generations, only one form exists, polymorphisms are gone, and there is monomorphism (fixation). Although the diagram shows alleles of one gene, this sort of fixation would be true of pretty much every gene locus or stretches of nucleotides. Even if the we had 1 million unfixed nucleotide loci, assuming the inbreeding didn’t kill the population, they would all fix pretty much in several generations just as this single gene got fixed to one allele from an original pool of 12.
The website cites an example where on one island all the individuals of a species were identical and probably followed this process:
in an introduced population (the island of Réunion, left), no polymorphisms are observed. This suggests that the founder population was very small, and that all variation has been lost.
In the original example, the 12 alleles came from pre-existing families, but let us suppose the 12 alleles came about from mutation. Suppose the 12 alleles came about by 1 mutation per gamete. The end result would be the same. But we can learn something from this.
First note, there is 1 mutation per each of the 12 gametes. We expect after several generations 1 mutant will get fixed and the other 11 mutant alleles lost to drift, but the bottom line is 1 mutant must necessarily be fixed in this forced illustration.
With a little imagination, we can extrapolate this forced illustration where I forced all the mutations onto the same gene locus to a more realistic situation where the mutations are spread to different loci. At each locus where a mutant allele might reside, any given mutant allele will have a 1 in 12 chance of going to fixation and an 11 in 12 chances of drifting into oblivion, but on average because there are 12 mutants each with a probability of 1/12 getting fixed, the expectation is 1 of the 12 mutants will get fixed and the other 11 go to extinction. The important point is, 1 mutant on average will get fixed because there was 1 mutation per gamete in generation 0.
And with yet a little more imagination, we can see if we increase the number of mutations per gamete in generation 0 to 100 mutation per gamete, we would on average get 100 new mutants fixed several generations down the line.
And yet with more imagination, if we were adding 100 mutations per gamete per generation, we’d be fixing 100 new mutants per generation. It just takes several generations to spool up to the point were fixation rates approach gametic mutation rates. With some math, it can be seen these ideas can be extrapolated to larger populations. But the bottom line is the fixation rate will approach the gametic mutation rate eventually for small well stirred populations. As Rupe and Sanford pointed out:
We show that neutral mutations go to fixation just as predicted by conventional theory (i.e., over deep time the fixation rate approached the gametic mutation rate).
The population sizes considered were 10,000 individuals.
But lost in all this is the quality of the mutants getting fixed. If the mutant is harmful but nearly neutral in terms of reproductive success, and if there are lots of these mutants, they will go to fixation! Thus the bad will just keep adding up, and the damage will be irreversible like a ratchet, hence we hear names like Muller’s and Haldane’s ratchets.
Part 2 describes the situation for small inbreeding well stirred populations. What happens in large non-inbreeding unstirred populations with lots of geographical isolation? That is the subject of the next post, but briefly, it doesn’t get necessarily better for large populations. I’ll cover that in part 3.
Discussion can also be pursued at CEU Forum: Neutral Evolution for Newbies.