Casey Luskin writes:
Despite disagreements, there is a standard story of human evolution that is retold in countless textbooks, news media articles, and documentaries. Indeed, virtually all the scientists I am citing here accept some evolutionary account of human origins, albeit flawed.
Starting with the early hominins and moving through the australopithecines, and then into the genus Homo, I will review the fossil evidence and assess whether it supports this standard account of human evolution. As we shall see, the evidence — or lack thereof — often contradicts this evolutionary story.
Early Hominins
In 2015, two leading paleoanthropologists reviewed the fossil evidence regarding human evolution in a prestigious scientific volume titled Macroevolution. They acknowledged the “dearth of unambiguous evidence for ancestor-descendant lineages,” and admitted,
[T]he evolutionary sequence for the majority of hominin lineages is unknown. Most hominin taxa, particularly early hominins, have no obvious ancestors, and in most cases ancestor-descendant sequences (fossil time series) cannot be reliably constructed.1
Nevertheless, numerous theories have been promoted about early hominins and their ancestral relationships to humans.
One leading fossil is described below:
Ardipithecus ramidus: Irish Stew or Breakthrough of the Year?
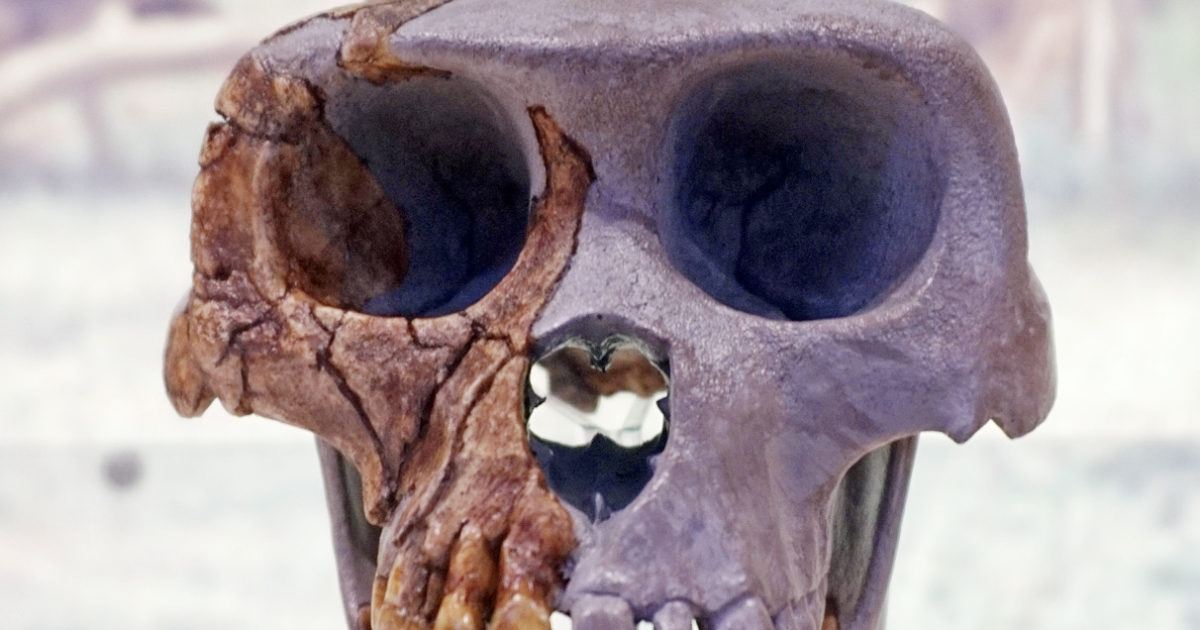
In 2009, Science announced the long-awaited publication of details about Ardipithecus ramidus (pictured above), a would-be hominin fossil that lived about 4.4 million years ago (mya). Expectations mounted after its discoverer, UC Berkeley paleoanthropologist Tim White, promised a “phenomenal individual” that would be the “Rosetta stone for understanding bipedalism.”17 The media eagerly employed the hominin they affectionately dubbed Ardi to evangelize the public for Darwin.
Discovery Channel ran the headline “‘Ardi,’ Oldest Human Ancestor, Unveiled,” and quoted White calling Ardi “as close as we have ever come to finding the last common ancestor of chimpanzees and humans.”18 The Associated Press declared, “World’s Oldest Human-Linked Skeleton Found,” and stated that “the new find provides evidence that chimps and humans evolved from some long-ago common ancestor.”19 Science named Ardi the “breakthrough of the year” for 2009,20 and introduced her with the headline, “A New Kind of Ancestor: Ardipithecus Unveiled.”21
Calling Ardi “new” may have been a poor word choice, for it was discovered in the early 1990s. Why did it take some 15 years to publish the analyses? A 2002 article in Science explains the bones were “soft,” “crushed,” “squished,” and “chalky.”22 Later reports similarly acknowledged that “portions of Ardi’s skeleton were found crushed nearly to smithereens and needed extensive digital reconstruction,” including the pelvis, which “looked like an Irish stew.”23
Claims about bipedal locomotion require accurate measurements of the precise shapes of key bones (like the pelvis). Can one trust declarations of a “Rosetta stone for understanding bipedalism” when Ardi was “crushed to smithereens”? Science quoted various paleoanthropologists who were “skeptical that the crushed pelvis really shows the anatomical details needed to demonstrate bipedality.”24
Even some who accepted Ardi’s reconstructions weren’t satisfied that the fossil was a bipedal human ancestor. Primatologist Esteban Sarmiento concluded in Science that “[a]ll of the Ar. ramidus bipedal characters cited also serve the mechanical requisites of quadrupedality, and in the case of Ar. ramidus foot-segment proportions, find their closest functional analog to those of gorillas, a terrestrial or semiterrestrial quadruped and not a facultative or habitual biped.”25 Bernard Wood questioned whether Ardi’s postcranial skeleton qualified it as a hominin,26 and co-wrote in Nature that if “Ardipithecus is assumed to be a hominin,” then it had “remarkably high levels of homoplasy [similarity] among extant great apes.”27 A 2021 study found that Ardi’s hands were well-suited for climbing and swinging in trees, and for knuckle-walking, giving it a chimp-like mode of locomotion.28 In other words, Ardi had ape-like characteristics which, if we set aside the preferences of Ardi’s promoters, should imply a closer relationship to apes than to humans. As the authors of the Nature article stated, Ardi’s “being a human ancestor is by no means the simplest, or most parsimonious explanation.”29Sarmiento even observed that Ardi had characteristics different from both humans and African apes, such as its unfused jaw joint, which ought to remove her far from human ancestry.30
Whatever Ardi was, everyone agrees the fossils was initially badly crushed and needed extensive reconstruction. No doubt this debate will continue, but are we obligated to accept the “human ancestor” position promoted by Ardi’s discoverers in the media? Sarmiento doesn’t think so. According Time magazine, he “regards the hype around Ardi to have been overblown.”31
Full article at Evolution News.
Notes
- Bernard Wood and Mark Grabowski, “Macroevolution in and around the Hominin Clade,” Macroevolution: Explanation, Interpretation, and Evidence, eds. Serrelli Emanuele and Nathalie Gontier (Heidelberg, Germany: Springer, 2015), 347-376.
- Michel Brunet et al., “Sahelanthropus or ‘Sahelpithecus’?,” Nature 419 (October 10, 2002), 582.
- Michel Brunet et al., “A new hominid from the Upper Miocene of Chad, Central Africa,” Nature 418 (July 11, 2002), 145-151. See also Michel Brunet et al., “New material of the earliest hominid from the Upper Miocene of Chad,” Nature 434 (April 7, 2005), 752-755.
- Smithsonian Natural Museum of Natural History, “Sahelanthropus tchadensis,” https://humanorigins.si.edu/evidence/human-fossils/species/sahelanthropus-tchadensis (accessed November 30, 2020).
- “Skull Find Sparks Controversy,” BBC News (July 12, 2002), http://news.bbc.co.uk/2/hi/science/nature/2125244.stm (accessed October 26, 2020).
- Milford Wolpoff et al., “Sahelanthropus or ‘Sahelpithecus’?” Nature 419 (October 10, 2002), 581-582.
- Roberto Macchiarelli et al., “Nature and relationships of Sahelanthropus tchadensis,” Journal of Human Evolution 149 (2020), 102898.
- Macchiarelli et al., “Nature and relationships of Sahelanthropus tchadensis.”
- Madelaine Böhme, quoted in Michael Marshall, “Our supposed earliest human relative may have walked on four legs,” New Scientist (November 18, 2020), https://www.newscientist.com/article/mg24833093-600-our-supposed-earliest-human-relative-may-have-walked-on-four-legs/ (accessed November 30, 2020).
- Bob Yirka, “Study of partial left femur suggests Sahelanthropus tchadensis was not a hominin after all,” Phys.org (November 24, 2020), https://phys.org/news/2020-11-partial-left-femur-sahelanthropus-tchadensis.html (accessed November 30, 2020).
- Potts and Sloan, What Does It Mean to Be Human?, 38.
- John Noble Wilford, “Fossils May Be Earliest Human Link,” New York Times (July 12, 2001), http://www.nytimes.com/2001/07/12/world/fossils-may-be-earliest-human-link.html (accessed October 26, 2020).
- John Noble Wilford, “On the Trail of a Few More Ancestors,” New York Times (April 8, 2001), http://www.nytimes.com/2001/04/08/world/on-the-trail-of-a-few-more-ancestors.html (accessed October 26, 2020).
- Leslie Aiello and Mark Collard, “Our Newest Oldest Ancestor?” Nature 410 (March 29, 2001), 526-527.
- K. Galik et al., “External and Internal Morphology of the BAR 1002’00 Orrorin tugenensis Femur,” Science 305 (September 3, 2004), 1450-1453.
- Sarmiento, Sawyer, and Milner, The Last Human, 35.
- Tim White, quoted in Ann Gibbons, “In Search of the First Hominids,” Science 295 (February 15, 2002), 1214-1219.
- Jennifer Viegas, “‘Ardi,’ Oldest Human Ancestor, Unveiled,” Discovery News (October 1, 2009), https://web.archive.org/web/20110613073934/http://news.discovery.com/history/ardi-human-ancestor.html (accessed October 26, 2020).
- Randolph Schmid, “World’s Oldest Human-Linked Skeleton Found,” NBC News (October 1, 2009), https://www.nbcnews.com/id/wbna33110809 (accessed October 26, 2020).
- Ann Gibbons, “Breakthrough of the Year: Ardipithecus ramidus,” Science 326 (December 18, 2009), 1598-1599.
- Gibbons, “New Kind of Ancestor,” 36-40.
- White, quoted in Gibbons, “In Search of the First Hominids,” 1214-1219, 1215-1216.
- Michael Lemonick and Andrea Dorfman, “Ardi Is a New Piece for the Evolution Puzzle,” Time (October 1, 2009), http://content.time.com/time/magazine/article/0,9171,1927289,00.html (accessed October 26, 2020).
- Gibbons, “New Kind of Ancestor,” 36-40, 39.
- Esteban Sarmiento, “Comment on the Paleobiology and Classification of Ardipithecus ramidus,” Science 328 (May 28, 2010), 1105b.
- Gibbons, “New Kind of Ancestor,” 36-40.
- Bernard Wood and Terry Harrison, “The Evolutionary Context of the First Hominins,” Nature 470 (February 17, 2011), 347-352.
- Thomas C. Prang, Kristen Ramirez, Mark Grabowski, and Scott A. Williams, “Ardipithecus hand provides evidence that humans and chimpanzees evolved from an ancestor with suspensory adaptations,” Science Advances 7 (February 24, 2021), eabf2474.
- New York University, “Fossils may look like human bones: Biological anthropologists question claims for human ancestry,” ScienceDaily (February 16, 2011), https://www.sciencedaily.com/releases/2011/02/110216132034.htm (accessed October 26, 2020).
- See Eben Harrell, “Ardi: The Human Ancestor Who Wasn’t?,” Time (May 27, 2010), http://content.time.com/time/health/article/0,8599,1992115,00.html (accessed October 26, 2020).
- Harrell, “Ardi: The Human Ancestor Who Wasn’t?”