The aim of this OP is to discuss in some order and with some completeness a few related objections to ID theory which are in a way connected to the argument that goes under the name of Texas Sharp Shooter Fallacy, sometimes used as a criticism of ID.
The argument that the TSS fallacy is a valid objection against ID has been many times presented by DNA_Jock, a very good discussant from the other side. So, I will refer in some detail to his arguments, as I understand them and remember them. Of course, if DNA_Jock thinks that I am misrepresenting his ideas, I am ready to ackowledge any correction about that. He can post here, if he can or likes, or at TSZ, where he is a contributor.
However, I thik that the issues discussed in this OP are of general interest, and that they touch some fundamental aspects of the debate.
As an help to those who read this, I will sum up the general structure of this OP, which will probably be rather long. I will discuss three different arguments, somewhat related. They are:
a) The application of the Texas Sharp Shooter fallacy to ID, and why that application is completely wrong.
b) The objection of the different possible levels of function definition.
c) The objection of the possible alternative solutions, and of the incomplete exploration of the search space.
Of course, the issue debated here is, as usual, the design inference, and in particular its application to biological objects.
So, let’s go.
a) The Texas Sharp Shooter fallacy and its wrong application to ID.
What’s the Texas Sharp Shooter fallacy (TSS)?
It is a logical fallacy. I quote here a brief description of the basic metaphor, from RationalWiki:
The fallacy’s name comes from a parable in which a Texan fires his gun at the side of a barn, paints a bullseye around the bullet hole, and claims to be a sharpshooter. Though the shot may have been totally random, he makes it appear as though he has performed a highly non-random act. In normal target practice, the bullseye defines a region of significance, and there’s a low probability of hitting it by firing in a random direction. However, when the region of significance is determined after the event has occurred, any outcome at all can be made to appear spectacularly improbable.
For our purposes, we will use a scenario where specific targets are apparently shot by a shooter. This is the scenario that best resembles what we see in biological objects, where we can observe a great number of functional structures, in particular proteins, and we try to understand the causes of their origin.
In ID, as well known, we use functional information as a measure of the improbability of an outcome. The general idea is similar to Paley’s argument for a watch: a very high level of specific functional information in an object is a very reliable marker of design.
But to evaluate functional information in any object, we must first define a function, because the measure of functional information depends on the function defined. And the observer must be free to define any possible function, and then measure the linked functional information. Respecting these premises, the idea is that if we observe any object that exhibits complex functional information (for example, more than 500 bits of functional information ) for an explicitly defined function (whatever it is) we can safely infer design.
Now, the objection that we are discussing here is that, according to some people (for example DNA_Jock), by defining the function after we have observed the object as we do in ID theory we are committing the TSS fallacy. I will show why that is not the case using an example, because examples are clearer than abstract words.
So, in our example, we have a shooter, a wall which is the target of the shooting, and the shootin itself. And we are the observers.
We know nothing of the shooter. But we know that a shooting takes place.
Our problem is:
- Is the shooting a random shooting? This is the null hypothesis
or:
- Is the shooter aiming at something? This is the “aiming” hypothesis
So, here I will use “aiming” instead of design, because my neo-darwinist readers will probably stay more relaxed. But, of course, aiming is a form of design (a conscious representation outputted to a material system).
Now I will describe three different scenarios, and I will deal in detail with the third.
-
First scenario: no fallacy.
In this case, we can look at the wall before the shooting. We see that there are 100 targets painted in different parts of the wall, rather randomly, with their beautiful colors (let’s say red and white). By the way, the wall is very big, so the targets are really a small part of the whole wall, even if taken together.
Then, we witness the shootin: 100 shots.
We go again to the wall, and we find that all 100 shots have hit the targets, one per target, and just at the center.
Without any worries, we infer aiming.
I will not compute the probabilities here, because we are not really interested in this scenario.
This is a good example of pre-definition of the function (the targets to be hit). I believe that neither DNA_Jock nor any other discussant will have problems here. This is not a TSS fallacy.
-
Second scenario: the fallacy.
The same setting as above. However, we cannot look at the wall before the shooting. No pre-specification.
After the shooting, we go to the wall and paint a target around each of the different shots, for a total of 100. Then we infer aiming.
Of course, this is exactly the TSS fallacy.
There is a post-hoc definition of the function. Moreover, the function is obviously built (painted) to correspond to the information in the shots (their location). More on this later.
Again, I will not deal in detail with this scenario because I suppose that we all agree: this is an example of TSS fallacy, and the aiming inference is wrong.
-
Third scenario: no fallacy.
The same setting as above. Again, we cannot look at the wall before the shooting. No pre-specification.
After the shooting, we go to the wall. This time, however, we don’t paint anything.
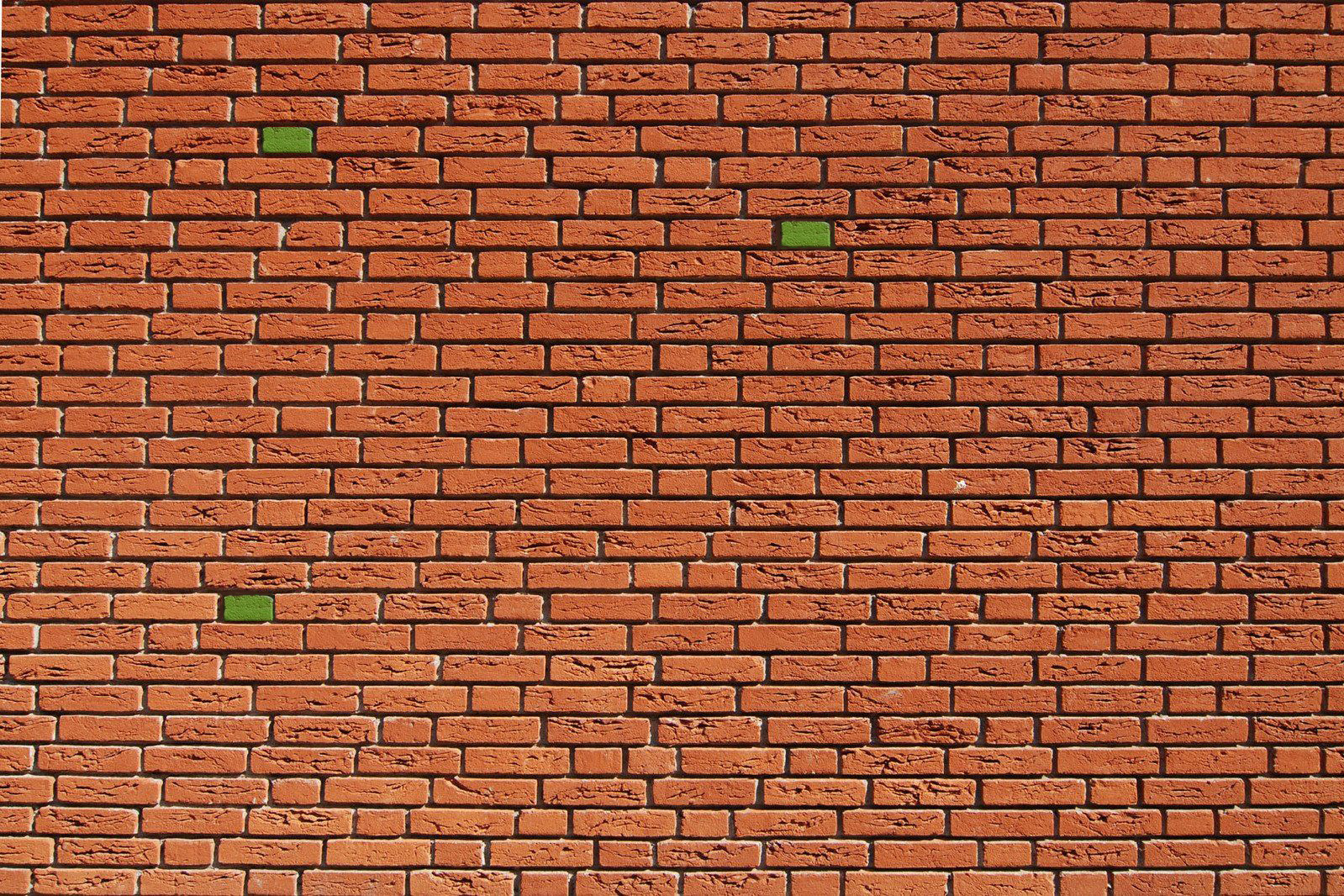
But we observe that the wall is made of bricks, small bricks. Almost all the bricks are brown. But there are a few that are green. Just a few. And they are randomly distributed in the wall.
We also observe that all the 100 shots have hit green bricks. No brown brick has been hit.
Then we infer aiming.
Of course, the inference is correct. No TSS fallacy here.
And yet, we are using a post-hoc definition of function: shooting the green bricks.
What’s the difference with the second scenario?
The difference is that the existence of the green bricks is not something we “paint”: it is an objective property of the wall. And, even if we do use something that we observe post-hoc (the fact that only the green bricks have been shot) to recognize the function post-hoc, we are not using in any way the information about the specific location of each shot to define the function. The function is defined objectively and independently from the contingent information about the shots.
IOWs, we are not saying: well the shooter was probably aiming at poin x1 (coordinates of the first shot) and point x2 (coordinates of the second shot), and so on. We just recognize that the shooter was aimin at the green bricks. An objective property of the wall.
IOWs ( I use many IOWs, because I know that this simple concept will meet a great resistance in the minds of our neo-darwinist friends) we are not “painting” the function, we are simply “recognizing” it, and using that recognition to define it.
Well, this third scenario is a good model of the design inference in ID. It corresponds very well to what we do in ID when we make a design inference for functional proteins. Therefore, the procedure we use in ID is no TSS fallacy. Not at all.
Given the importance of this model for our discussion, I will try to make it more quantitative.
Let’s say that the wall is made of 10,000 bricks in total.
Let’s say that there are only 100 green bricks, randomly distributed in the wall.
Let’s say that all the green bricks have been hit, and no brown brick.
What are the probabilities of that result if the null hypothesis is true (IOWs, if the shooter was not aiming at anything) ?
The probability of one succesful hit (where success means hitting a green brick) is of course 0.01 (100/10000).
The probability of having 100 successes in 100 shots can be computed using the binomial distribution. It is:
10e-200
IOWs, the system exhibits 664 bits of functional information. More ore less like the TRIM62 protein, an E3 ligase discussed in my previous OP about the Ubiquitin system, which exhibits an increase of 681 bits of human conserved functional information at the transition to vertebrates.
Now, let’s stop for a moment for a very important step. I am asking all neo-darwinists who are reading this OP a very simple question:
In the above situation, do you infer aiming?
It’s very important, so I will ask it a second time, a little louder:
In the above situation, do you infer aiming?
Because if your answer is no, if you still think that the above scenario is a case of TSS fallacy, if you still believe that the observed result is not unlikely, that it is perfectly reasonable under the assumption of a random shooting, then you can stop here: you can stop reading this OP, you can stop discussing ID, at least with me. I will go on with the discussion with the reasonable people who are left.
So, in the end of this section, let’s remind once more the truth about post-hoc definitions:
- No post-hoc definition of the function that “paints” the function using the information from the specific details of what is observed is correct. Those definitions are clear examples of TSS fallacy.
- On the contrary, any post-hoc definition that simply recognizes a function which is related to an objectively existing property of the system, and makes no special use of the specific details of what is observed to “paint” the function, is perfectly correct. It is not a case of TSS fallacy.
b) The objection of the different possible levels of function definition.
DNA_Jock summed up this specific objection in the course of a long discussion in the thread about the English language:
Well, I have yet to see an IDist come up with a post-specification that wasn’t a fallacy. Let’s just say that you have to be really, really, really cautious if you are applying a post-facto specification to an event that you have already observed, and then trying to calculate how unlikely that specific event was. You can make the probability arbitrarily small by making the specification arbitrarily precise.
OK, I have just discussed why post-specifications are not in themselves a fallacy. Let’s say that DNA_Jock apparently admits it, because he just says that we have to be very cautious in applying them. I agree with that, and I have explained what the caution should be about.
Of course, I don’t agree that ID’s post-hoc specifications are a fallacy. They are not, not at all.
And I absolutely don’t agree with his argument that one of the reasosn why ID’s post-hoc specifications are a fallacy would be that “You can make the probability arbitrarily small by making the specification arbitrarily precise.”
Let’s try to understand why.
So, let’s go back to our example 3), the wall with the green bricks and the aiming inference.
Let’s make our shooter a little less precise: let’s say that, out of 100 shots, only 50 hits are green bricks.
Now, the math becomes:
The probability of one succesful hit (where success means hitting a green brick) is still 0.01 (100/10000).
The probability of having 50 successes or more in 100 shots can be computed using the binomial distribution. It is:
6.165016e-72
Now, the system exhibits “only” 236 bits of functional information. Much less than in the previous example, but still more than enough, IMO, to infer aiming.
Consider that five sigma, which is ofetn used as a standard in physics to reject the nulll hypothesis , is just 3×10-7, less than 22 bits.
Now, DNA_Jock’s objection would be that our post-hoc specification is not valid because “we can make the probability arbitrarily small by making the specification arbitrarily precise”.
But is that true? Of course not.
Let’s say that, in this case, we try to “make the specification arbitrarily more precise”, defining the function of sharp aiming as “hitting only green bricks with all 100 shots”.
Well, we are definitely “making the probability arbitrarily small by making the specification arbitrarily precise”. Indeed, we are making the specification more precise for about 128 orders of magnitude! How smart we are, aren’t we?
But if we do that, what happens?
A very simple thing: the facts that we are observing do not meet the specification anymore!
Because, of course, the shooter hit only 50 green bricks out of 100. He is smart, but not that smart.
Neither are we smart if we do such a foolish thing, defining a function that is not met by observed facts!
The simple truth is: we cannot at all “make the probability arbitrarily small by making the specification arbitrarily precise”, as DNA_Jock argues, in our post-hoc specification, because otherwise our facts would not meet our specification anymore, and that would be completely useless and irrelevant..
What we can and must do is exactly what is always done in all cases where hypothesis testing is applied in science (and believe me, that happens very often).
We compute the probabilities of observing the effect that we are indeed observing, or a higher one, if we assume the null hypothesis.
That’s why I have said that the probability of “having 50 successes or more in 100 shots” is 6.165016e-72.
This is called a tail probability, in particular the probability of the upper tail. And it’s exactly what is done in science, in most scenarios.
Therefore, DNA_Jock’s argument is completely wrong.
c) The objection of the possible alternative solutions, and of the incomplete exploration of the search space.
c1) The premise
This is certainly the most complex point, because it depends critically on our understanding of protein functional space, which is far from complete.
For the discussion to be in some way complete, I have to present first a very general premise. Neo-darwinists, or at least the best of them, when they understand that they have nothing better to say, usually desperately recur to a set of arguments related to the functional space of proteins. The reason is simple enough: as the nature and structure of that space is still not well known or understood, it’s easier to equivocate with false reasonings.
Their purpose, in the end, is always to suggest that functional sequences can be much more frequent than we believe. Or at least, that they are much more frequent than IDists believe. Because, if functional sequences are frequent, it’s certainly easier for RV to find them.
The arguments for this imaginary frequency of biological function are essentially of five kinds:
- The definition of biological function.
- The idea that there are a lot of functional islands.
- The idea that functional islands are big.
- The idea that functional islands are connected. The extreme form of this argument is that functional islands simply don’t exist.
- The idea that the proteins we are observing are only optimized forms that derive from simpler implementations through some naturally selectable ladder of simple steps.
Of course, different mixtures of the above arguments are also frequently used.
OK. let’s get rid of the first, which is rather easy. Of course, if we define extremely simple biological functions, they will be relatively frequent.
For example, the famous Szostak experiment shows that a weak affinity for ATP is relatively common in a random library; about 1 in 1011 sequences 80 AAs long.
A weak affinity for ATP is certainly a valid definition for a biological function. But it is a function which is at the same time irrelevant and non naturally selectable. Only naturally selectable functions have any interest for the neo-darwinian theory.
Moreover, most biological functions that we observe in proteins are extremely complex. A lot of them have a functional complexity beyond 500 bits.
So, we are only interested in functions in the protein space which are naturally selectable, and we are specially interested in functions that are complex, because those are the ones about which we make a design inference.
The other three points are subtler.
- The idea that there are a lot of functional islands.
Of course, we don’t know exactly how many functional islands exist in the protein space, even restricting the concept of function to what was said above. Neo-darwinists hope that there are a lot of them. I think there are many, but not so many.
But the problem, again, is drastically redimensioned if we consider that not all functional islands will do. Going back to point 1, we need naturally selectable islands. And what can be naturally selected is much less than what can potentially be functional. A naturally selectable island of function must be able to give a reproductive advantage. In a system that already has some high complexity, like any living cell, the number of functions that can be immediately integrated in what already exists, is certainly strongly constrained.
This point is also stricly connected to the other two points, so I will go on with them and then try some synthesis.
- The idea that functional islands are big.
Of course, functional islands can be of very different sizes. That depends on how many sequences, related at sequence level (IOWs, that are part of the same island), can implement the function.
Measuring functional information in a sequence by conservation, like in the Dustron method or in my procedure many times described, is an indirect way of measuring the size of a functional island. The greater is the functional complexity of an island, the smaller is its size in the search space.
Now, we must remember a few things. Let’s take as an example an extremely conserved but not too long sequence, our friend ubiquitin. It’s 76 AAs long. Therefore, the associated search space is 20^76: 328 bits.
Of course, even the ubiquitin sequence can tolerate some variation, but it is still one of the most conserved sequences in evolutionary history. Let’s say, for simplicity, that at least 70 AAs are stictly conserved, and that 6 can vary freely (of course, that’s not exact, just an approximation for the sake of our discussion).
Therefore, using the absolute information potential of 4.3 bits per aminoacid, we have:
Functional information in the sequence = 303 bits
Size of the functional island = 328 – 303 = 25 bits
Now, a functional island of 25 bits is not exactly small: it corresponds to about 33.5 million sequences.
But it is infinitely tiny if compared to the search space of 328 bits: 7.5 x 10^98 sequences!
If the sequence is longer, the relationship between island space and search space (the ocean where the island is placed) becomes much worse.
The beta chain of ATP synthase (529 AAs), another old friend, exhibits 334 identities between e. coli and humans. Always for the sake of simplicity, let’s consider that about 300 AAs are strictly conserved, and let’s ignore the functional contraint on all the other AA sites. That gives us:
Search space = 20^529 = 2286 bits
Functional information in the sequence = 1297 bits
Size of the functional island = 2286 – 1297 = 989 bits
So, with this computation, there could be about 10^297 sequences that can implement the function of the beta chain of ATP synthase. That seems a huge number (indeed, it’s definitley an overestimate, but I always try to be generous, especially when discussing a very general principle). However, now the functional island is 10^390 times smaller than the ocean, while in the case of ubiquitin it was “just” 10^91 times smaller.
IOWs, the search space (the ocean) increases exponentially much more quickly than the target space (the functional island) as the lenght of the functional sequence increases, provided of course that the sequences always retain high functional information.
The important point is not the absolute size of the island, but its rate to the vastness of the ocean.
So, the beta chain of ATP synthase is really a tiny, tiny island, much smaller than ubiquitin.
Now, what would be a big island? It’s simple: a functional isalnd which can implement the same function at the same level, but with low functional information. The lower the functional information, the bigger the island.
Are there big islands? For simple functions, certainly yes. Behe quotes the antifreeze protein as an example example. It has rather low FI.
But are there big islands for complex functions, like that of ATP synthase beta chain? It’s absolutely reasonable to believe that there are none. Because the function here is very complex, and it cannot be implemented by a simple sequence, exactly like a functional spreadsheet software annot be written by a few bits of source code. Neo-darwinists will say that we don’t know that for certain. It’s true, we don’t know it for certain. We know it almost for certain.
The simple fact remains: the only example of the beta chain of the F1 complex of ATP synthase that we know of is extremely complex.
Let’s go, for the moment, to the 4th argument.
- The idea that functional islands are connected. The extreme form of this argument is that functional islands simply don’t exist.
This is easier. We have a lot of evidence that functional islands are not connected, and that they are indeed islands, widely isolated in the search space of possible sequences. I will mention the two best evidences:
4a) All the functional proteins that we know of, those that exist in all the proteomse we have examined, are grouped in abot 2000 superfamilies. By definition, a protein superfamily is a cluster of sequences that have:
- no sequence similarity
- no structure similarity
- no function similarity
with all the other groups.
IOWs, islands in the sequence space.
4b) The best (and probably the only) good paper that relates an experiment where Natural Selection is really tested by an approrpiaite simulation is the rugged landscape paper:
Experimental Rugged Fitness Landscape in Protein Sequence Space
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0000096
Here, NS is correctly simulated in a phage system, because what is measured is infectivity, which in phages is of course strictly related to fitness.
The function studied is the retrieval of a partially damaged infectivity due to a partial random substitution in a protein linked to infectivity.
In brief, the results show a rugged landscape of protein function, where random variation and NS can rather easily find some low-level peaks of function, while the original wild-type, optimal peak of function cannot realistically be found, not only in the lab simulation, but in any realistic natural setting. I quote from the conclusions:
The question remains regarding how large a population is required to reach the fitness of the wild-type phage. The relative fitness of the wild-type phage, or rather the native D2 domain, is almost equivalent to the global peak of the fitness landscape. By extrapolation, we estimated that adaptive walking requires a library size of 1070 with 35 substitutions to reach comparable fitness.
I would recommend to have a look at Fig. 5 in the paper to have an idea of what a rugged landscape is.
However, I will happily accept a suggestion from DNA_Jock, made in one of his recent comments at TSZ about my Ubiquitin thread, and with which I fully agree. I quote him:
To understand exploration one, we have to rely on in vitro evolution experiments such as Hayashi et al 2006 and Keefe & Szostak, 2001. The former also demonstrates that explorations one and two are quite different. Gpuccio is aware of this: in fact it was he who provided me with the link to Hayashi – see here.
You may have heard of hill-climbing algorithms. Personally, I prefer my landscapes inverted, for the simple reason that, absent a barrier, a population will inexorably roll downhill to greater fitness. So when you ask:How did it get into this optimized condition which shows a highly specified AA sequence?
I reply
It fell there. And now it is stuck in a crevice that tells you nothing about the surface whence it came. Your design inference is unsupported.
Of course, I don’t agree with the last phrase. But I fully agree that we should think of local optima as “holes”, and not as “peaks”. That is the correct way.
So, the protein landscape is more like a ball and holes game, but without a guiding labyrinth: as long as the ball in on the flat plane (non functional sequences), it can go in any direction, freely. However, when it falls into a hole, it will quickly go to the bottom, and most likely it will remain there.
But:
- The holes are rare, and they are of different sizes
- They are distant from one another
- A same function can be implemented by different, distant holes, of different size
What does the rugged landscape paper tell us?
- That the wildtype function that we observe in nature is an extremely small hole. To find it by RV and NS, according to the authors, we should start with a library of 10^70 sequences.
- That there are other bigger holes which can partially implement some function retrieval, and that are in the range of reasonable RV + NS
- That those simpler solutions are not bridges to the optimal solution observed in the wildtype. IOWs. they are different, and there is no “ladder” that NS can use to reach the optimal solution .
Indeed, falling into a bigger hole (a much bigger hole, indeed) is rather a severe obstacle to finding the tiny hole of the wildtype. Finding it is already almost impossible because it is so tiny, but it becomes even more impossible if the ball falls into a big hole, because it will be trapped there by NS.
Therefore, to sum up, both the existence of 2000 isolated protein superfamilies and the evidence from the rugged landscape paper demonstrate that functional islands exist, and that they are isolated in the sequence space.
Let’s go now to the 5th argument:
- The idea that the proteins we are observing are only optimized forms that derive from simpler implementations by a naturally selectable ladder.
This is derived from the previous argument. If bigger functional holes do exist for a function (IOWs, simpler implementations), and they are definitely easier to find than the optimal solution we observe, why not believe that the simpler solutions were found first, and then opened the way to the optimal solution by a process of gradual optimization and natural selection of the steps? IOWs, a naturally selectable ladder?
And the answer is: because that is impossible, and all the evidence we have is against that idea.
First of all, even if we know that simpler implementations do exist in some cases (see the rugged landscape paper), it is not at all obvious that they exist as a general rule.
Indeed, the rugged landscape experiment is a very special case, because it is about retrieval of a function that has been only partially impaired by substituting a random sequence to part of an already existing, functional protein.
The reason for that is that, if they had completely knocked out the protein, infectivity, and therefore survival itself, would not have survived, and NS could not have acted at all.
In function retrieval cases, where the function is however kept even if at a reduced level, the role of NS is greatly helped: the function is already there, and can be optimed with a few naturally selectable steps.
And that is what happens in the case of the Hayashi paper. But the function is retrieved only very partially, and, as the authors say, there is no reasonable way to find the wildtype sequence, the optimal sequence, in that way. Because the optimal sequence would require, according to the authors, 35 AA substitutions, and a starting library of 10^70 random sequences.
What is equally important is that the holes found in the experiment are not connected to the optimal solution (the wildtype). They are different from it at sequence level.
IOWs, this bigger holes do not lead to the optimal solution. Not at all.
So, we have a strange situation: 2000 protein superfamilies, and thousand and tousands of proteins in them, that appear to be, in most cases, extremely functional, probably absolutely optimal. But we have absolutely no evidence that they have been “optimized”. They are optimal, but not necessarily optimized.
Now, I am not excluding that some optimization can take place in non design systems: we have good examples of that in the few known microevolutionary cases. But that optimization is always extremely short, just a few AAs substitutions once the starting functional island has been found, and the function must already be there.
So, let’s say that if the extremely tiny functional island where our optimal solution lies, for example the wildtype island in the rugged landscape experiment, can be found in some way, then some small optimization inside that functional island could certainly take place.
But first, we have to find that island: and for that we need 35 specific AA substitutions (about 180 bits), and 10^70 starting sequences, if we go by RV + NS. Practically impossible.
But there is more. Do those simpler solutions always exist? I will argue that it is not so in the general case.
For example, in the case of the alpha and beta chains of the F1 subunit of ATP synthase, there is no evidence at all that simpler solutions exist. More on that later.
So, to sum it up:
The ocean of the search space, according to the reasonings of neo-darwinists, should be overflowing with potential naturally selectable functions. This is not true, but let’s assume for a moment, for the sake of discussion, that it is.
But, as we have seen, simpler functions or solutions, when they exist, are much bigger functional islands than the extremely tiny functional islands corresponding to solutions with high functional complexity.
And yet, we have seen that there is absolutely no evidence that simpler solutuion, when they exist, are bridges, or ladder, to highly complex solutions. Indeed, there is good evidence of the contrary.
Given those premises, what would you expect if the neo-darwinian scenario were true? It’s rather simple: an universal proteome overflowing with simple functional solutions.
Instead, what do we observe? It’s rather simple: an universal proteome overflowing with highly functional, probably optimal, solutions.
IOWs, we find in the existing proteome almost exclusively highly complex solutions, and not simple solutions.
The obvious conclusion? The neo-darwinist scenario is false. The highly functional, optimal solutions that we observe can only be the result of intentional and intelligent design.
c2) DNA_Jock’s arguments
Now I will take in more detail DNA_Jock’ s two arguments about alternative solutions and the partial exploration of the protein space, and will explain why they are only variants of what I have already discussed, and therefore not valid.
The first argument, that we can call “the existence of alternative solutions”, can be traced to this statement by DNA_Jock:
Every time an IDist comes along and claims that THIS protein, with THIS degree of constraint, is the ONLY way to achieve [function of interest], subsequent events prove them wrong. OMagain enjoys laughing about “the” bacterial flagellum; John Walker and Praveen Nina laugh about “the” ATPase; Anthony Keefe and Jack Szostak laugh about ATP-binding; now Corneel and I are laughing about ubiquitin ligase: multiple ligases can ubiquinate a given target, therefore the IDist assumption is false. The different ligases that share targets ARE “other peaks”.
This is Texas Sharp Shooter.
We will debate the laugh later. For the moment, let’s see what the argument states.
It says: the solution we are observing is not the only one. There can be others, in some cases we know there are others. Therefore, your computation of probabilities, and therefore of functional inpormation, is wrong.
Another way to put it is to ask the question: “how many needles are there in the haystack?”
Alan Fox seems to prefer this metaphor:
This is what is wrong with “Islands-of-function” arguments. We don’t know how many needles are in the haystack. G Puccio doesn’t know how many needles are in the haystack. Evolution doesn’t need to search exhaustively, just stumble on a useful needle.
They both seem to agree about the “stumbling”. DNA_Jock says:
So when you ask:
How did it get into this optimized condition which shows a highly specified AA sequence?
I reply
It fell there. And now it is stuck in a crevice that tells you nothing about the surface whence it came.
OK, I think the idea is clear enough. It is essentially the same idea as in point 2 of my general premise. There are many functional islands. In particular, in this form, many functional islands for the same function.
I will answer it in two parts:
- Is it true that the existence of alternative solutions, if they exist, makes the computation of functional complexity wrong?
- Have we really evidence that alternative solutions exist, and of how frequent they can really be?
I will discuss the first part here, and say something about the second part later in the OP.
Let’s read again the essence of the argument, as summed up by me above:
” The solution we are observing is not the only one. There can be others, in some cases we know there are others. Therefore, your computation of probabilities, and therefore of functional information, is wrong.”
As it happens with smart arguments (and DNA_Jock is usually smart), it contains some truth, but is essentially wrong.
The truth could be stated as follows:
” The solution we are observing is not the only one. There can be others, in some cases we know there are others. Therefore, our computation of probabilities, and therefore of functional information, is not completely precise, but it is essentially correct”.
To see why that is the case, let’s use again a very good metaphor: Paley’s old watch. That will help to clarify my argument, and then I will discuss how it relies to proteins, in particular.
So, we have a watch. Whose function is to measure time. And, in general, let’s assume that we infer design for the watch, because its functional information is high enough to exclude that it could appear in any non design system spontaneously. I am confident that all reasonable people will agree with that. Anyway, we are assuming it for the present discussion.
Now, after having made a design inference (a perfectly correct inference, I would say) for this object, we have a sudden doubt. We ask ourselves: what if DNA_Jock is right?
So, we wonder: are there other solutions to measure time? Are there other functional islands in the search space of material objects?
Of course there are.
I will just mention four clear examples: a sundial, an hourglass, a digital clock, an atomic clock.
The sundial uses the position of the sun. The hourglass uses a trickle of sand. The digital clock uses an electronic oscillator that is regulated by a quartz crystal to keep time. An atomic clock uses an electron transition frequency in the microwave, optical, or ultraviolet region.
None of them uses gears or springs.
Now, two important points:
- Even if the functional complexity of the five above mentioned solutions is probably rather different (the sundial and the hourglass are probably quite simpler, and the atomic clock is probably the most complex), they are all rather complex. None of them would be easily explained without a design inference. IOWs, they are small functional islands, each of them. Some are bigger, some are really tiny, but none of them is big enough to allow a random origin in a non design system.
- None of the four additional solutions mentioned would be, in any way, a starting point to get to the traditional watch by small functional modifications. Why? Because they are completely different solutions, based on different ideas and plans.
If someone believes differently, he can try to explain in some detail how we can get to a traditional watch starting from an hourglass.
Now, an important question:
Does the existence of the four mentioned alternative solutions, or maybe of other possible similar solutions, make the design inference for the traditional watch less correct?
The answer, of course, is no.
But why?
It’s simple. Let’s say, just for the sake of discussion, that the traditional watch has a functional complexity of 600 bits. There are at least 4 additional solutions. Let’s say that each of them has, again, a functional complexity of 500 bits.
How much does that change the probability of getting the watch?
The answer is: 2 bits (because we have 4 solutions instead of one). So, now the probability is 598 bits.
But, of course, there can be many more solutions. Let’s say 1000. Now the probability would be about 590 bits. Let’s say one million different complex solutions (this is becoming generous, I would say). 580 bits. One billion? 570 bits.
Shall I go on?
When the search space is really huge, the number of really complex solutions is empirically irrelevant to the design inference. One observed complex solution is more than enough to infer design. Correctly.
We could call this argument: “How many needles do you need to tranfsorm a haystack into a needlestack?” And the answer is: really a lot of them.
Our poor 4 alternative solutions will not do the trick.
But what if there are a number of functional islands that are much bigger, much more likely? Let’s say 50 bits functional islands. Much simpler solutions. Let’s say 4 of them. That would make the scenario more credible. Not so much, probably, but certainly it would work better than the 4 complex solutions.
OK, I have already discussed that above, but let’s say it again. Let’s say that you have 4 (or more) 50 bits solution, and one (or more) 500 bits solution. But what you observe as a fact is the 500 bits solution, and none of the 50 bits solutions. Is that credible?
No, it isn’t. Do you know how smaller a 500 bits solution is if compared to a 50 bits solution? It’s 2^450 times smaller: 10^135 times smaller. We are dealing with exponential values here.
So, if much simpler solutions existed, we would expect to observe one of them, and not certainly a solution that is 10^135 times more unlikely. The design inference for the highly complex solution is not disturbed in any way by the existence of much simpler solutions.
OK, I think that the idea is clear enough.
c3) The laughs
As already mentioned, the issue of alternative solutions and uncounted needles seems to be a special source of hilarity for DNA_Jock. Good for him (a laugh is always a good thing for physical and mental health). But are the laughs justified?
I quote here again his comment about the laughs, that I will use to analyze the issues.
Every time an IDist comes along and claims that THIS protein, with THIS degree of constraint, is the ONLY way to achieve [function of interest], subsequent events prove them wrong. OMagain enjoys laughing about “the” bacterial flagellum; John Walker and Praveen Nina laugh about “the” ATPase; Anthony Keefe and Jack Szostak laugh about ATP-binding; now Corneel and I are laughing about ubiquitin ligase: multiple ligases can ubiquinate a given target, therefore the IDist assumption is false. The different ligases that share targets ARE “other peaks”.
I will not consider the bacterial flagellum, that has no direct relevance to the discussion here. I will analyze, instead, the other three laughable issues:
- Szostak and Keefe’s ATP binding protein
- ATP synthase (rather than ATPase)
- E3 ligases
Szostak and Keefe should not laugh at all, if they ever did. I have already discussed their paper a lot of times. It’s a paper about directed evolution which generates a strongly ATP binding protein form a weakly ATP binding protein present in a random library. It is directed evolution by mutation and artificial selection. The important point is that both the original weakly binding protein and the final strongly binding protein are not naturally selectable.
Indeed, a protein that just binds ATP is of course of no utility in a cellular context. Evidence of this obvious fact can be found here:
A Man-Made ATP-Binding Protein Evolved Independent of Nature Causes Abnormal Growth in Bacterial Cells
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0007385
There is nothing to laugh about here: the protein is a designed protein, and anyway it is no functional peak/hole at all in the sequence space, because it cannot be naturally selected.
Let’s go to ATP synthase.
DNA_Jock had already remarked:
They make a second error (as Entropy noted) when they fail to consider non-traditional ATPases (Nina et al).
And he gives the following link:
Highly Divergent Mitochondrial ATP Synthase Complexes in Tetrahymena thermophila
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2903591/
And, of course, he laughs with Nina (supposedly).
OK. I have already discussed that the existence of one or more highly functional, but different, solutions to ATP building would not change the ID inference at all. But is it really true that there are these other solutions?
Yes and no.
As far as my personal argument is concerned, the answer is definitely no (or at least, there is no evidence of them). Why?
Because my argument, repeated for years, has always been based (everyone can check) on the alpha and beta chains of ATP synthase, the main constituents of the F1 subunit, where the true catalytic function is implemented.
To be clear, ATP synthase is a very complex molecule, made of many different chains and of two main multiprotein subunits. I have always discussed only the alpha and beta chains, because those are the chains that are really highly conserved, from prokaryotes to humans.
The other chains are rather conserved too, but much less. So, I have never used them for my argument. I have never presented blast values regarding the other chains, or made any inference about them. This can be checked by everyone.
Now, the Nina paper is about a different solution for ATP synthase that can be found in some single celled eukaryotes,
I quote here the first part of the abstract:
The F-type ATP synthase complex is a rotary nano-motor driven by proton motive force to synthesize ATP. Its F1 sector catalyzes ATP synthesis, whereas the Fo sector conducts the protons and provides a stator for the rotary action of the complex. Components of both F1 and Fo sectors are highly conserved across prokaryotes and eukaryotes. Therefore, it was a surprise that genes encoding the a and b subunits as well as other components of the Fo sector were undetectable in the sequenced genomes of a variety of apicomplexan parasites. While the parasitic existence of these organisms could explain the apparent incomplete nature of ATP synthase in Apicomplexa, genes for these essential components were absent even in Tetrahymena thermophila, a free-living ciliate belonging to a sister clade of Apicomplexa, which demonstrates robust oxidative phosphorylation. This observation raises the possibility that the entire clade of Alveolata may have invented novel means to operate ATP synthase complexes.
Emphasis mine.
As everyone can see, it is absolutely true that these protists have a different, alternative form of ATP symthase: it is based on a similar, but certainly divergent, architecture, and it uses some completely different chains. Which is certainly very interesting.
But this difference does not involve the sequence of the alpha and beta chains in the F1 subunit.
Beware, the a and b subunits mentioned above by the paper are not the alpha and beta chains.
From the paper:
The results revealed that Spot 1, and to a lesser extent, spot 3 contained conventional ATP synthase subunits including α, β, γ, OSCP, and c (ATP9)
IOWs, the “different” ATP synthase uses the same “conventional” forms of alpha and beta chain.
To be sure of that, I have, as usual, blasted them against the human forms. Here are the results:
ATP synthase subunit alpha, Tetrahymena thermophila, (546 AAs) Uniprot Q24HY8, vs ATP synthase subunit alpha, Homo sapiens, 553 AAs (P25705)
Bitscore: 558 bits Identities: 285 Positives: 371
ATP synthase subunit beta, Tetrahymena thermophila, (497 AAs) Uniprot I7LZV1, vs ATP synthase subunit beta, Homo sapiens, 529 AAs (P06576)
Bitscore: 729 bits Identities: 357 Positives: 408
These are the same, old, conventional sequences that we find in all organisms, the only sequences that I have ever used for my argument.
Therefore, for these two fundamental sequences, we have no evidence at all of any alternative peaks/holes. Which, if they existed, would however be irrelevant, as already discussed.
Not much to laugh about.
Finally, E3 ligases. DNA_Jock is ready to laugh about them because of this very good paper:
Systematic approaches to identify E3 ligase substrates
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5103871/
His idea, shared with other TSZ guys, is that the paper demonstrates that E3 ligases are not specific proteins, because a same substrate can bind to more than one E3 ligase.
The paper says:
Significant degrees of redundancy and multiplicity. Any particular substrate may be targeted by multiple E3 ligases at different sites, and a single E3 ligase may target multiple substrates under different conditions or in different cellular compartments. This drives a huge diversity in spatial and temporal control of ubiquitylation (reviewed by ref. [61]). Cellular context is an important consideration, as substrate–ligase pairs identified by biochemical methods may not be expressed or interact in the same sub-cellular compartment.
I have already commented elsewhere (in the Ubiquitin thread) that the fact that a substrate can be targeted by multiple E3 ligases at different sites, or in different sub-cellular compartments, is clear evidence of complex specificity. IOWs, its’ not that two or more E3 ligases bind a same target just to do the same thing, they bind the same target in different ways and different context to do different things. The paper, even if very interesting, is only about detecting affinities, not function.
That should be enough to stop the laughs. However, I will add another simple concept. If E3 ligases were really redundant in the sense suggested by DNA_Jock and friends, their loss of function should not be a serious problem for us. OK, I will just quote a few papers (not many, because this OP is already long enough):
The multifaceted role of the E3 ubiquitin ligase HOIL-1: beyond linear ubiquitination.
https://www.ncbi.nlm.nih.gov/pubmed/26085217
HOIL-1 has been linked with antiviral signaling, iron and xenobiotic metabolism, cell death, and cancer. HOIL-1 deficiency in humans leads to myopathy, amylopectinosis, auto-inflammation, and immunodeficiency associated with an increased frequency of bacterial infections.
WWP1: a versatile ubiquitin E3 ligase in signaling and diseases.
https://www.ncbi.nlm.nih.gov/pubmed/22051607
WWP1 has been implicated in several diseases, such as cancers, infectious diseases, neurological diseases, and aging.
RING domain E3 ubiquitin ligases.
https://www.ncbi.nlm.nih.gov/pubmed/19489725
RING-based E3s are specified by over 600 human genes, surpassing the 518 protein kinase genes. Accordingly, RING E3s have been linked to the control of many cellular processes and to multiple human diseases. Despite their critical importance, our knowledge of the physiological partners, biological functions, substrates, and mechanism of action for most RING E3s remains at a rudimentary stage.
HECT-type E3 ubiquitin ligases in nerve cell development and synapse physiology.
https://www.ncbi.nlm.nih.gov/pubmed/25979171
The development of neurons is precisely controlled. Nerve cells are born from progenitor cells, migrate to their future target sites, extend dendrites and an axon to form synapses, and thus establish neural networks. All these processes are governed by multiple intracellular signaling cascades, among which ubiquitylation has emerged as a potent regulatory principle that determines protein function and turnover. Dysfunctions of E3 ubiquitin ligases or aberrant ubiquitin signaling contribute to a variety of brain disorders like X-linked mental retardation, schizophrenia, autism or Parkinson’s disease. In this review, we summarize recent findings about molecular pathways that involve E3 ligasesof the Homologous to E6-AP C-terminus (HECT) family and that control neuritogenesis, neuronal polarity formation, and synaptic transmission.
Finally I would highly recommend the following recent paper to all who want to approach seriously the problem of specificity in the ubiquitin system:
Specificity and disease in the ubiquitin system
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5264512/
Abstract
Post-translational modification (PTM) of proteins by ubiquitination is an essential cellular regulatory process. Such regulation drives the cell cycle and cell division, signalling and secretory pathways, DNA replication and repair processes and protein quality control and degradation pathways. A huge range of ubiquitin signals can be generated depending on the specificity and catalytic activity of the enzymes required for attachment of ubiquitin to a given target. As a consequence of its importance to eukaryotic life, dysfunction in the ubiquitin system leads to many disease states, including cancers and neurodegeneration. This review takes a retrospective look at our progress in understanding the molecular mechanisms that govern the specificity of ubiquitin conjugation.
Concluding remarks
Our studies show that achieving specificity within a given pathway can be established by specific interactions between the enzymatic components of the conjugation machinery, as seen in the exclusive FANCL–Ube2T interaction. By contrast, where a broad spectrum of modifications is required, this can be achieved through association of the conjugation machinery with the common denominator, ubiquitin, as seen in the case of Parkin. There are many outstanding questions to understanding the mechanisms governing substrate selection and lysine targeting. Importantly, we do not yet understand what makes a particular lysine and/or a particular substrate a good target for ubiquitination. Subunits and co-activators of the APC/C multi-subunit E3 ligase complex recognize short, conserved motifs (D [221] and KEN [222] boxes) on substrates leading to their ubiquitination [223–225]. Interactions between the RING and E2 subunits reduce the available radius for substrate lysines in the case of a disordered substrate [226]. Rbx1, a RING protein integral to cullin-RING ligases, supports neddylation of Cullin-1 via a substrate-driven optimization of the catalytic machinery [227], whereas in the case of HECT E3 ligases, conformational changes within the E3 itself determine lysine selection [97]. However, when it comes to specific targets such as FANCI and FANCD2, how the essential lysine is targeted is unclear. Does this specificity rely on interactions between FA proteins? Are there inhibitory interactions that prevent modification of nearby lysines? One notable absence in our understanding of ubiquitin signalling is a ‘consensus’ ubiquitination motif. Large-scale proteomic analyses of ubiquitination sites have revealed the extent of this challenge, with seemingly no lysine discrimination at the primary sequence level in the case of the CRLs [228]. Furthermore, the apparent promiscuity of Parkin suggests the possibility that ubiquitinated proteins are the primary target of Parkin activity. It is likely that multiple structures of specific and promiscuous ligases in action will be required to understand substrate specificity in full.
To conclude, a few words about the issue of the sequence space not entirely traversed.
We have 2000 protein superfamilies that are completely unrelated at sequence level. That is evidence that functional protein sequences are not bound to any particular region of the sequence space.
Moreover, neutral variation in non coding and non functional sequences can go any direction, without any specific functional constraints. I suppose that neo-darwinists would recognize that parts of the genomes is non functional, wouldn’t they? And we have already seen elsewhere (in the ubiquitin thread discussion) that many new genes arise from non coding sequences.
So, there is no reason to believe that the functional space has not been traversed. But, of course, neutral variation can traverse it only at very low resolution.
IOWs, there is no reason that any specific part of the sequence space is hidden from RV. But of course, the low probabilistic resources of RV can only traverse different parts of the sequence space occasionally.
It’s like having a few balls that can move freely on a plane, and occasionally fall into a hole. If the balls are really few and the plane is extremely big, the balls will be able to potentially traverse all the regions of the plane, but they will pass only through a very limited number of possible trajectories. That’s why finding a very small hole will be almost impossible, wherever it is. And there is no reason to believe that small functional holes are not scattered in the sequence space, as protein superfamilies clearly show.
So, it’s not true that highly functional proteins are hidden in some unexplored tresure trove in the sequence space. They are there for anyone to find them, in different and distant parts of the sequence space, but it is almost impossible to find them through a random walk, because they are so small.
And yet, 2000 highly functional superfamilies are there.
Moreover, The rate of appearance of new suprefamilies is highest at the beginning of natural history (for example in LUCA), when a smaller part of the sequence space is likely to have been traversed, and decreases constantly, becoming extremely low in the last hundreds of million years. That’s not what you would expect if the problem of finding new functional islands were due to how much sequence space has been traversed, and if the sequence space were really so overflowing with potential naturally selectable functions, as neo-darwinists like to believe.
OK, that’s enough. As expected, this OP is very long. However, I think that it was important to discuss all these partially related issues in the same context.