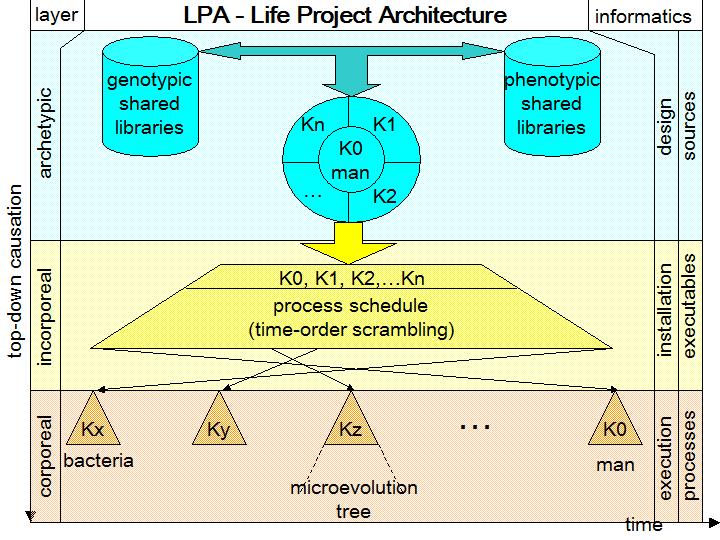
A point that Darwinists make is that anti-Darwinists have not developed any theory for the origins of species, and think that is a weakness. But for an IDer/creationist is not so difficult to have ideas about solutions of the problem of origins. I for one developed a proposal for a theory, and I will illustrate it here. I called it “LPA” (Life Project Architecture). See below its simple schema, where the x-axis is time and the y-axis the top-down intelligent causation:
LPA model for origins is cent percent design based. The role of natural selection, about the creation of biological information and complexity, is null. To grasp LPA one must entirely invert the reasoning of evolutionism. This means in the same time to assume a solid informatics engineering perspective and apply it to biology.
An axiom of Darwinism is the corporeal descent of all species from a simple unique corporeal common ancestor, by means of unintelligent processes. To pass to LPA this axiom has to be somehow inverted and becomes: abstract descent of all kinds from a unique complex abstract common archetype-kind, in the context of a global integrated project of life on Earth. While in Darwinism all the core stuff happens only at the corporeal level, in LPA it happens at the incorporeal level. The inversion of LPA compared to Darwinism not only substitutes non intelligent causes with intelligence but considers two layers upon the corporeal layer, the only one considered by materialist evolutionism. Darwinism is one-dimensional, LPA is three-dimensional.
This inversion of LPA vs. Darwinism (which in a sense is a restoration of truth) immediately removes the major absurdities and problems of evolutionism:
(1) Intelligence, able to create information and organization, substitutes chance and necessity, which are unable.
(2) The “less” comes from the “more” and not the opposite, as in evolutionism.
(3) The corporeal macroevolution, which is a real “engineering suicide”, is substituted with an abstract incorporeal derivation, done at a higher layer than the material one. The bodies have no more to materially and monstrously morph, with all the countless complications one can imagine.
(4) The Darwinian material common ancestor of minimal complexity becomes the immaterial template of the more complex being, from which all the simpler beings abstractly derive as sub-projects, when some major characteristics and architectural plans are changed.
(5) What is intelligent is created by intelligence. It is obvious that the unintelligent cannot produce the intelligent, as evolutionism pretends.
(6) The incapability of chance and necessity to create semiotic processing, sophisticated molecular machinery, CSI, correlated CSI, irreducible complexity, functional hierarchies, etc.
Who is the more complex living being that can pretend to be the “maximum project template” in LPA? Answer: man. Before this answer Darwinists object that man arose last in history, then cannot be first. In LPA man can well be the first in the order of design and last in the order of time. Conversely, bacteria, which are the simplest organisms, are last in the order of design and first in the time order. In fact, you can see in the middle of the schema, at the incorporeal layer, the possibility of a total scrambling between design order and time order. This is the flexibility that informatics shows so well. By the way, it is indeed the appearance over time from the simplest to the more complex species that caused the illusion of an evolution from simplicity to complexity, while instead it was simply a specific order of execution of a complexity already fully present from the beginning in the design incorporeal layer.
The LPA explains also all homologies, analogies and similarities between all living beings, at the genetic, morphological, functional and system levels, because LPA implements very strong “common design” “shared libraries” “high level integration” “software reuse” paradigms. In fact, in the schema, at the top level, you see the shared libraries (genotypic and phenotypic) strongly interrelated each other and in turn with the core design, by means of a tri-way bus/channel.
LPA entails three layers: archetypical, incorporeal and corporeal (on the left side of the schema). They are related in the informatics jargon (on the right side) to: design, installation and deployment, execution. The design phase deals with the “sources”, the installation phase deals with “executables” and the execution phase involves “processes” and outputs. Any computer programmer will recognize the standard iter of informatics engineering. LPA could quite well represent how an informatics or robotics engineer, in charge of developing a series of different robots, would organize the overall project, according to a high-level integrated vision. Warning: here I don’t affirm that living beings are simple robots. They are far higher and more complex than robots. But this, to greater reason, reinforces the ID argument, because this implies that their design and construction must be far more sophisticated than modern robotics.
The shift of the job from matter to abstractness, which greatly increases the flexibility, is a fundamental engineering principle, which LPA implements. The history of engineering and technology shows us that engineers have always strived to shift their job from matter to mind. Today engineers use high level abstract tools (CAE, CAD, CAM, etc.). No engineer would use directly low level Darwinian material evolution. Darwinism is a decrepit bankrupt way of thinking, unable to construct the least system. ID LPA is a modern and technologically sound way to construct complex systems.
Another point about LPA is that it agrees with engineering and in the same time is consistent with traditional cosmogony (when is correctly interpreted), according to which the manifestation of macrocosms and microcosms (which are analogous and share the same layering) is a top-down causation ID process that starts from archetypical principles, passes trough an intermediate incorporeal formation and finally concludes into the corporeal world. In LPA the teaching of tradition about creation and engineering meet. Not by chance or by my invention, rather because both describe complex constructions of intelligence. All sciences of construction necessarily share some basic principles that are universal. LPA meets such principles.
Also about the position of man in nature LPA is inverted respecting evolutionism because is “man-centric”. Here again tradition and engineering meet. In any traditional doctrine man is the “imago-Dei” “center of creation”, the hierarchically higher being who synthesizes in himself all others lower peripheral beings. Analogously, if an engineer wants to construct – say – a computer, he starts to think about the central processing unit, then he will develop the peripheral devices.
An important point is why I called the living beings as “kinds”, K0, K1, K2… in the LPA model. The high level design at the archetype layer doesn’t need to specify in advance and in details all the countless species, variants and races of a kind. These variants will differ from the parent kind only in some minor details, while kinds differ in major architectural body plan aspects. What is sufficient is to carefully design and insert into a set of main kinds the complex potentialities needed to generate their minor variants during reproduction of populations in history, by means of endogen and hexogen triggering signals or factors at run time. In few words, the biological “software” must be highly flexible and parametrized, scalable, allowing plasticity and micro-evolvability. I called “microevolution trees” the trees of variants for each kind K and I symbolized them with triangles inside the final corporeal layer. I believe it is sensible to identify the Ks to what in the classic taxonomy are families/kinds. Also most creationists agree on that. As example, Nathaniel T. Jeanson writes “reproductive compatibility identifies membership within a kind. Breeding experiments identify the classification rank of family (kingdom-phylum-class-order-family-genus-species) as roughly defining the boundaries of each kind”. This level is roughly the threshold below which microevolution trees begin.
What evidences support LPA? To my knowledge, the main biological data known so far – if you dismiss the evolutionist eyeglasses – fit well with LPA, because common design explains all things that Darwinian common descent seems to explain. With the difference that LPA has not the absurdities and contradictions that Darwinism has.