Fig. 1: Cnidaria are probably one of the most ancient phyla of known Metazoa.
Metazoa, or multicellular organisms, are one of the amazing “novelties” in natural history. At some point, single eukaryotic cells begin to be organized in a new, incredibly complex plan: a multicellular organism.
It is well known and understood that one of the main tools to realize that innovation, that new expression of life, is cell differentiation. Cells, while sharing the same genome, become different, incredibly different one from the other. Stem cells differentiate and acquire, through amazing and still poorly understood epigenetic trajectories, completely different cellular phenotypes and functions. The miracle of transcription regulation, as we have seen in another recent OP, is at the center of multiple levels of control involved in that achievement.
But there is another important aspect in metazoa that is even less understood, and at least equally amazing. Cells do not only differentiate: they must form tissues and organs and, in the end, a whole body plan. Body plans are the distinctive feature of the highest categorization of metazoa, the phyla, those individualized general programs that, strangely, appear almost all at the same time at the famous Cambrian Explosion (or, in different and lost forms, even earlier, at the Ediacaran Explosion).
But body plans are made of individual organs, and individual organs are made of different tissues. How can cells, while differentiating and implementing their specific epigenetic programs, form tissues and organs and organisms?
First of all, we should ask: where are cells, while they do that? The subject of this OP is about that question: the relationship of individual cells with their “environment” in the organisms that is being formed and built.
Let’s clarify immediately a few points:
- The environment that we are considering here is not really an outer environment. IOWs, it is not the outer world, with all its contingent varieties. It is, instead, an environment that surrounds the cells and is strictly controlled and regulated. It is called the Extracellular Matrix (ECM).
- The ECM is created by none other than the cells themselves. They generate it, they control it, they regulate it, they remodel it in many different and complex ways. As we will see.
- The ECM is the environment where cells live, differentiate and move. It controls and regulates the individual cells in many ways. As we will see.
- The main feature that contributes to generate tissues, organs and body plans, together with cell differentiation, is cell migration. Cells have to move, to migrate according to a precise and strictly regulated plan. Without controlled migration no tissue, no organ can be formed. Cells that differentiate differently must migrate to different places in the growing organism.
- Cell migration is not only a pillar of organism development. It remains a basic feature of many cell systems even in “adult” life. That is specially true for important cell systems, first of all the immune system.
- All cell migrations happen in the ECM, and are controlled and implemented by the complex interactions between the cells and the ECM.
- Those interactions, surprisingly, are not only biochemical (as we usually imagine cell interactions to be), but mainly mechanical.
- The cell structures that mediate those interactions between the cells and their ECM are one of the most amazing examples of functional complexity in the cell. Yes, like the spliceosome, the ubiquitin system and the transcription regulation networks, that we have considered previously. As we will see
So, this is a brief summary, just to help the reader to follow the many aspects of this topic that will now be presented in greater detail.
Sounds interesting? It is.
-
The Extracellular Matrix (ECM)
The ECM is, very simply, the 3D environment where cells exist. It is a very complex structure, and it is very different in different organisms and in different tissues. In bones, the ECM is strongly mineralized, in chartilage it is very dense but not mineralized. In other connective tissues, like reticular tissues, it can be very loose. For circulating cells in blood, plasma is the ECM. A special type of ECM is the basal membrane, which supports epitelial cells.
Here are two good reviews about ECM:
The extracellular matrix at a glance
Biology of the Extracellular Matrix: An Overview
This is how the second paper starts:
The extracellular matrix (ECM) is an intricate network composed of an array of multidomain macromolecules organized in a cell/tissue-specific manner. Components of the ECM link together to form a structurally stable composite, contributing to the mechanical properties of tissues. The ECM is also a reservoir of growth factors and bioactive molecules. It is a highly dynamic entity that is of vital importance, determining and controlling the most fundamental behaviors and characteristics of cells such as proliferation, adhesion, migration, polarity, differentiation, and apoptosis.
So, let’s understand better.
The cells that form a multicellular organism are embedded in a self-generated semi-fluid environment, the ECM. All of them.
Of course, some types of cell are also in contact with the external environment, the world. For example, epithelial cells represent the outer surface of the organism to the external world, for example the epidermis. But the basal part of the epithelial tissue is anyway firmly rooted in some form of ECM.
The ECM is, first of all, water with soluble components in it. This fluid part is called Extracellular fluid, or also Interstitial fluid. That water is our internal ocean, where the basic exhanges between cells and the rest of the organism (nutrients, oxygen, and so on) take place. While this fluid component is very important, it is not really the subject of our discussion here.
In this fluid component, however, cells secrete a lot of important insoluble big proteins and other molecules, that make the ECM what it is, in different tissues. Those substances are “immersed” in the Extracellular fluid, and determine the physical and mechanical properties of that outer environment.
Here is a good general review of those components of ECM, the so called matrisome:
Overview of the Matrisome—An Inventory of Extracellular Matrix Constituents and Functions
While there are many components of the ECM and the matrisome, they can be essentially categorized in three rather different classes of proteins:
- Collagens
- Glicoproteins
- Proteoglicans
About 1000 proteins seem to be implied in the composition of the ECM. However, almost 300 seem to be part of a basic core matrisome. I will refer here mainly to a recent compilation of that core matrisome (see in particular Fig. 1):
The extracellular matrix: Tools and insights for the “omics” era
Collagens
Collagens are fibrous proteins that build the basic structure of the ECM in most tissues.
I quote from the paper “Overview of the Matrisome”:
Collagens are found in all metazoa and provide structural strength to all forms of extracellular matrices, including the strong fibers of tendons, the organic matrices of bones and cartilages, the laminar sheets of basement membranes, the viscous matrix of the vitreous humor, and the interstitial ECMs of the dermis and of capsules around organs.
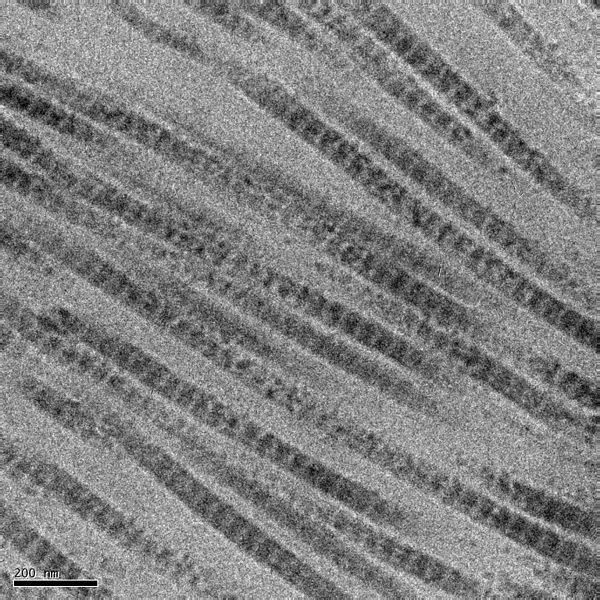
Fig. 2: Vincent R. Sherman and Maria I. Lopez. Transmission electron micrograph of the collagen fibrils in rabbit skin. Licensed under the Creative Commons Attribution-Share Alike 4.0 International license.
There are about 28 different collagen types, assembled from 44 different molecules, according to the core matrisome described in the second paper quoted above.
Collagens form the hard structure of the ECM. To simplify, each basic molecule of collagen (procollagen) is made of three collagen chains (for example, type 1 collagen is made of 2 alpha-1 chains and 1 alpha-2 chain), that form a right-handed triple helix. Out of the cell, those procollagen molecules are assemble into long fibrils and fibers.
One important concept is that, from the point of view os sequence, collagen is a relatively “simple” molecule, because it is formed mainly by highly repetitive short sequences of three AAs one of which is usually glycine, while proline and hydrossyproline are highly represented too. So, this is a rather “repetitive” sequence. But even so, the many modifications of the pattern generate an amazing complexity of the final result: 44 different genes and basic sequences are assembled into 28 different types of collagen triple helics, which have high specific properties and very different expression in different tissues. So, type I collagen is the most common form, present in skin, tendons, bones and many other important tissues. Type II is rather specific of chartilage. Type III makes the delicate reticular fibers so important in loose ECM. Type IV is mainly represented in the extremely important basal lamina that makes the basement membran in epithelial tissues. And so on.
Glycoproteins
These are proteins which contain oligosaccharide chains. There are 195 of them included in the essential core matrisome mentioned above. Some of them are exremely important. In particular:
- Elastin, which confers elasticity to tissues.
- Laminin, which is an imprtant component of basal lamina in the basement membrane.
- Fibronectin, about which we have to spend a few more words.
Fibronectin is a protein, or better a group of proteins, that connects to the other components of the ECM (collagens, proteoglycans) and to the cell (by the integrins, see later). Therefore, it is central in the implementation of practically all the complex functions in cell-ECM communication.
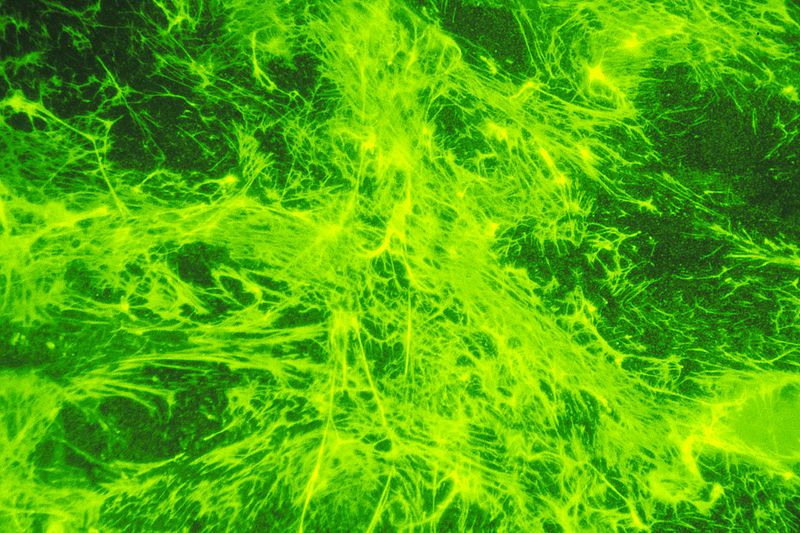
Fig. 3: Fluorescent-labeled fibronectin on a layer of cells. It appears to be green in color. Fibronectin, a protein glue that anchors cells and hold them together often disappears when cells become cancerous. https://commons.wikimedia.org/wiki/File:Fibronectin_(1).jpg Source: National Cancer Institute Author: Linda Bartlett (Photographer) [Public domain or Public domain], via Wikimedia Commons
Here is a very good review about fibronectin:
Assembly of Fibronectin Extracellular Matrix
And here are a few key points from that paper:
The extracellular matrix (ECM) has been recognized as an essential structural component in multicellular organisms for millennia (Plato trans. 1965). However, the old view of ECM as an inert scaffold is clearly incorrect. ECM is a dynamic network, a reservoir for growth factors and fluids, and an essential organizer of tissues, cellular microenvironments, and stem cell niches. It shows exquisite tissue specificity and adapts to changes in age, development, and disease. Even so, the molecular events that assemble secreted ECM proteins into complex networks are still not completely understood.
***
Fibronectin (FN) is a ubiquitous ECM glycoprotein that is assembled into a fibrillar matrix in all tissues and throughout all stages of life. Its assembly is a cell-mediated process (McDonald 1988) and is essential for life (George et al. 1993). FN fibrils form linear and branched meshworks around cells and connect neighboring cells
***
FN is a modular protein composed of types I, II, and III repeating units (Figure 1). Two intramolecular disulfide bonds form within each type I and type II module to stabilize the folded structure. Type III modules are seven-stranded β-barrel structures that lack disulfides (Leahy et al. 1996, Potts & Campbell 1994). Modules are organized into binding sites for collagen/gelatin, integrins, heparin, FN, and other extracellular molecules (Figure 1).
***
FN exists in multiple isoforms generated by alternative splicing. The single FN gene transcript encodes 12 isoforms in rodents and cows and 20 isoforms in humans.
Fibronectin is a 2386 AAs long protein (in humans). As said, it is still a highly modular and partially repetitive sequence, but at a much more sophisticated level than collagen. Moreover, it definitely exhibits a huge re-engineering in vertebrates, with an information jump of 0.92 bits per aminoacid, corresponding to 2188 bits. With the necessary caution implied by the partially repetitive structure of the sequence, I would definitely say that those are huge numbers.
The “fibronectin meshwork” is therefore involved in many functions, but the one we will be focusing on here is its role, in association with integrins, in mechanosensing and cell migration.
Proteoglycans
Proteoglycans, previously known as mucopolysaccharides, are heavily glycosilated proteins, where the non protein content is represented by one or more chain of Glycosaminoglycan (GAG, a class of hetero-polysaccharides), and amounts to 50-60% of the molecule by weight (vs only 10-15% in glycoproteins).
They are important components of the ECM too. There are 35 of them listed in the essential core matrisome quoted above.
They have been classified according to the GAG present in the molecule: Hyaluronates, Dermatan sulfates, Chondroitin sulfates, Heparan sulfates, Keratan sulfates. But the recent classifications are based on the protein component, and include: Aggrecan, Neurocan, Syndecans, Versican and many others.
They are not only found in ECM: they are important components of the cell membrane, too.
Their most evident role is mechanical: together with collagen, extracellular proteoglycans are the main determinants of the mechanical properties of ECM, like stiffness, elasticity, compressibility, and so on. But they have also a lot of biochemical functions. The topic is very complex, so for the moment we will not deal with it in detail.
-
The cell-matrix interaction: Integrins and the Adhesome
How does the cell interact with the ECM?
There are many different ways, indeed. But we will focus here on the main and best known: the system of integrins.
Integrins are a family of transmembrane proteins that have a fundamental role in connecting cells to the ECM. They are heterodimers, and they are always made of an alpha chain + a beta chain. In mammals there are at least 18 alpha chains and at least 8 beta chains, forming at least 24 different heterodimers. Different heterodimers have different specificities and roles.
Alpha chains are about 1000 AAs long, beta chains are about 800 AAs long. While different alpha chains share moderate homology (for example, 25% identity between alpha 1 and alpha 5), and so do beta chains (for example, 46% identity between beta 1 and beta 2), alpha and beta chains share practically no homology between them.
Integrins are exquisitely metazoan proteins. A very simple repertoire is present in the simplest Metazoa, but the full repertoire that we find in mammals is detectable only in chordates, and more specifically in vertebrates. See here:
Integrins: Bidirectional Allosteric Signaling Machines
The Integrin Receptor Family: Evolution and Complexity
Integrins are restricted to the metazoa; no homologs are detected in prokaryotes, plants, or fungi (Whittaker and Hynes, 2002). The simplest metazoa, sponges and cnidaria, have integrins (Burke 1999, Hughes 2001) and it is clear that primitive bilateria had at least two integrin αβ heterodimers, the descendents of which persist to this day in organisms as diverse as flies, nematodes, and vertebrates (Hynes and Zhao, 2000). Indeed, that is the entire set of integrins in Caenorhabditis elegans; one β subunit and two α subunits forming two integrins. Orthologs of these two integrins are recognized in Drosophila melanogaster and in vertebrates, although vertebrates have expanded each set (Figure 1). One set (blue in Figure 1) recognizes the tripeptide sequence, RGD, in molecules such as fibronectin and vitronectin in vertebrates and tiggrin in Drosophila, whereas the other set (purple in Figure 1) mediates adhesion to basement membrane laminins. It is plausible that evolution of integrins was necessary to allow the cell-matrix adhesion intrinsic to metazoa, and as diploblastic organisms evolved, the two cell layers may have evolved separate integrins to mediate their asymmetric interactions with the basal lamina; representatives of these two primordial integrins are detected in all higher metazoan phyla.
Each of the 24 integrins shown in Figure 1 appears to have a specific, nonredundant function. In part, this is evident from the details of their ligand specificities (not shown in Figure 1) but is most clearly shown by the phenotypes of knockout mice (Table 1). Genes for the β subunits and all but four of the α subunits have been knocked out and each phenotype is distinct, reflecting the different roles of the various integrins. The phenotypes range from a complete block in preimplantation development (β1), through major developmental defects (α4, α5, αv, β8), to perinatal lethality (α3, α6, α8, αv, β4, β8) and defects in leukocyte function (αL, αM, αE, β2, β7), inflammation (β6), hemostasis (αIIb, β3, α2), bone remodeling (β3), and angiogenesis (α1, β3) as well as others
As transmembrane proteins, Integrins bind ECM components (ligands) with their extracellular part, and inner components of the cell, in particular actin, with their intracellular part. Both these interations are very complex. For those interested in details, I would recommend the following very good site, MBINFO, realized by the Mechanobiology Institute of the University of Singapore. I will use a few of their very clear images in the following discussion.
a) Integrin – ECM interaction.
The extracellular domain of the Integrin involves both chains, alpha and beta, and represents the bigger part of the molecule. Both chains have a transmebrane part, and a smaller intracellular tail.
The general structure is common to all integrin molecules. In a very simplified way, both the alpha and the beta chain can be described as a big extracellular “head” which rests on a very long “leg” that can bend at a “knee” region, and then goes through the membrane and ends with the intracellular tail.
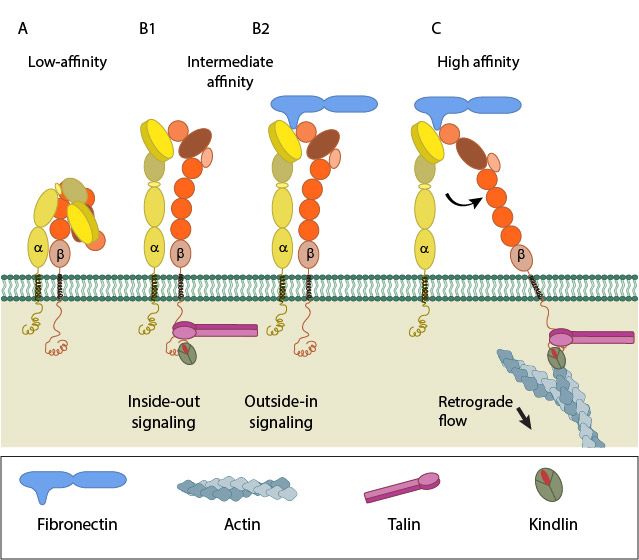
Fig 4: A. Low-affinity integrin has an inactive, bent, conformation. B1 and B2. Inside-out integrin activation by cytoplasmic proteins or Outside-in integrin activation via ECM ligands both lead to complete extension of the extracellular domains. C. The hallmark of open, high-affinity activated integrin is separation of the cytoplasmic leg domains. From: “How is integrin activated?” licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
Essentially, the Integrin can exist in two basic conformations:
a) The inactive conformation (A in Fig.1), in which the two chains are bent at the “knee”, and the intracellular tails are very near.
b) A fully active conformation, usually induced by interaction with inner components (B) or with extracellular ligands in the ECM (C), in which the two “legs” become fully extended, and divaricated, so that the two intracellular tails are separated (D). The extracellular ligand interacts with the two “heads” in the open configuration.
Here is a similar, but more detailed, image of the two basic configurations:
Fig. 5: Inactive conformation of integrin (left) and a hypothetical model of the open, active form (right), with a fibrinogen peptide in red and a talin domain in magenta. From: PDB-101 Molecule of the month: Integrin. Attribution: David S. Goodsell and the RCSB PDB. Licensed under a CC-BY-4.0 license
Following activation, integrins tend to migrate in the cell membrane and to form dynamic clusterings. Those clusterings of activated integrins mature into highly dynamic structures called Focal adhesions (FA).
We will see more about the complex functions of FAs later. For the moment, let’s anticipate that FAs are the key complex structure that mediates cell-ECM interactions, and that they have at least two major functions:
- In stationary cells, they anchor the cell to the surroundung ECM, and help to make the cell morphology stable.
- They are the essential structures that allow cell migration.
Fig. 6 shows a mature Focal Adhesion complex:
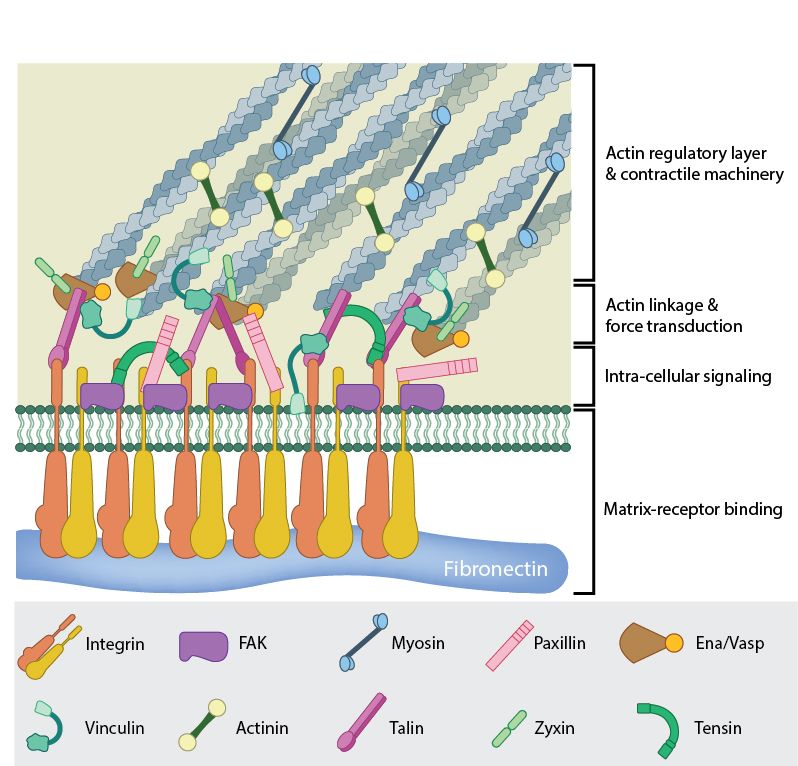
Fig 6: A mature FA contains hundreds of proteins that are grouped based on their contribution to four basic processes: receptor/matrix binding, linkage to actin cytoskeleton, intracellular signal transduction, and actin polymerization. Both actin polymerization and actomyosin contractile machinery generate forces that affect mechanosensitive proteins in the actin linking module, the receptor module (e.g. integrins), the signaling module, and the actin polymerization module. The combined activity of the mechanosensitive components form the mechanoresponsive network. The theoretical organization and protein-protein interactions as shown are based upon references.
So, to sum it up, integrins connect the cell to the ECM by:
a) Passing from the inactive conformation to the active conformation (“legs” extended, open active site between the two “heads”), in response to intracellular or extracellular signals.
b) Binding some specific ligand in some specific component of he ECM (for example, fibronectin).
c) Migrating in the cell membrane to form specific clusterings and, in the end, a very complex structure called Focal Adhesion.
A very important point is that FAs, even if very complex, are extremely dynamic: they arise as “nascent” FAs, then grow into mature FAs, and finally they are quickly disassembled.
b) Integrin – cytoskeleton interaction: the Adhesome.
The other side of the Integrin molecule, and therefore of the Focal Adhesion, is of course the link to the cell sytoskeleton, in particular to actin filaments.
Very briefly, actin is a family of globular proteins (G-actin) that polymerize in the form of micro-filaments (F-actin). Those actin filaments form a very dynamic cytoskeleton scaffold in the cell. Together with myosin, they can form contractile structures.
Now, we could think that at least the connection of the inner tails of Integrins to the actin cytoskeleton should be relatively simple. Well, that’s absolutely not the case.
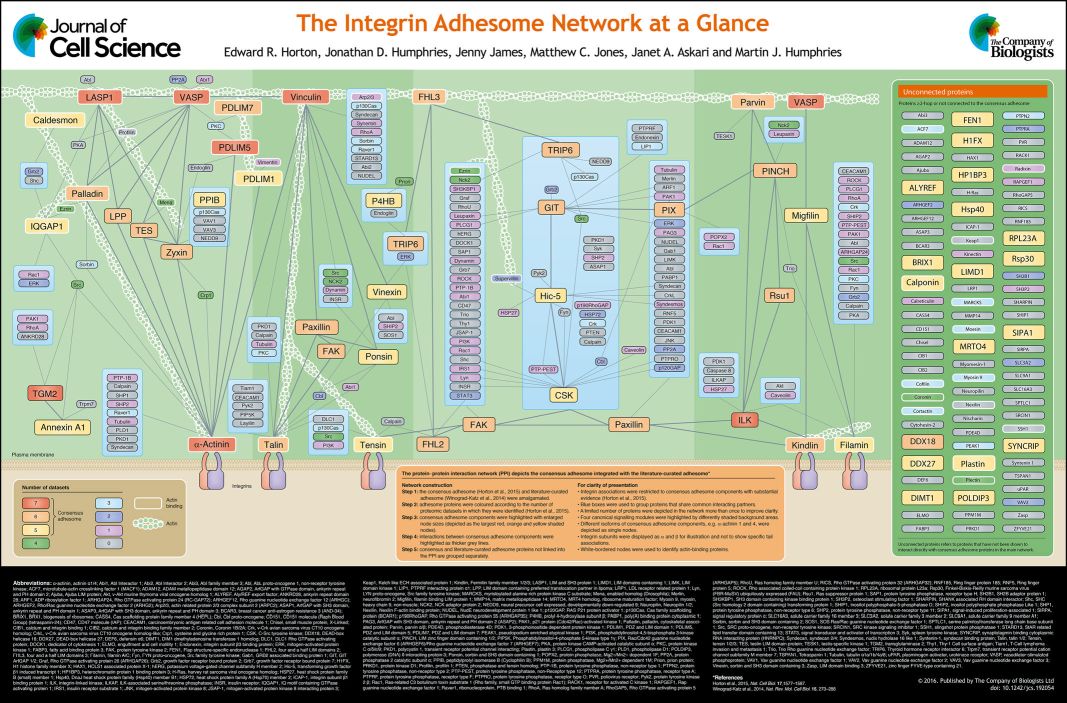
Integrins do not connect directly to actin. They are connected by an extremely complex structure, that is usually called the Adhesome, or Integrin Adhesome.
The Adhesome is an amazing multiprotein structure that assemles dynamically at the intracellular side of Integrins, completing and stabilizing the Focal Adhesion.
Here is a summary table of its known components:
Fig. 7: The integrin adhesome network at a glance. From The integrin adhesome network at a glance , licensed under CC BY 3.0
There is also a web-site completely dedicated to the Integrin Adhesome. Here it is:
The Adhesome: A Focal Adhesion Network
The site lists at present 150 “bona fide components” of the Adhesome + 82 associated components, and a total of 6,542 interactions. Not bad!
Some important points about the Adhesome:
a) Like the FA, of which it is the main component, it is extremely dynamic. The lifetime of individual proteins in the Adhesome is in the order of seconds, while the whole FA has a mean lifetime in the order of tens of minutes.
b) The proteins in the Adhesome are usually sensitive to mechanical force, and recruited by it (we will come back to that later)
c) While many of the functions and structures in the Adhesome are still poorly understood, it seems clear that it is higly organized in space, with a structure that involves three different functional layers: an Integrin Signaling Layer (nearest to the cell membrane), an intermediate Force Transduction Layer and an inner Actin Regulatory Layer, each with its specific protein compositions. See Fig. 2 here:
The “Stressful” Life of Cell Adhesion Molecules: On the Mechanosensitivity of Integrin Adhesome
Of course, we cannot detail here the 150+ proteins that assemble in the Adhesome. We will just describe in some detail the most important and best studied of them: talin.
c) Talin
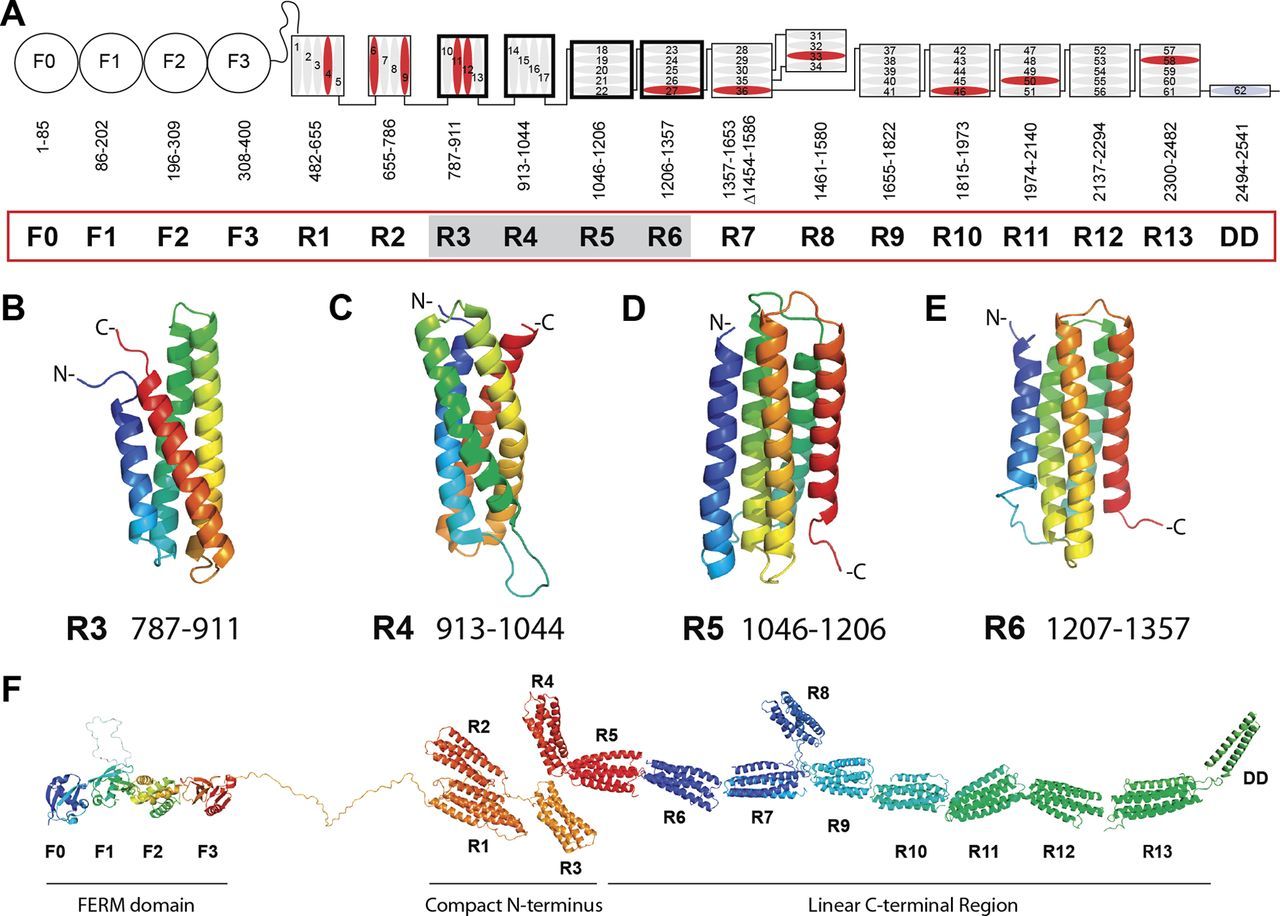
Talin is a 2500+ AAs protein which implements a central and extremely complex role in Focal Adhesions. Its structure includes an N-terminal “head” (a FERM domain made of 4 subunits, F0-F3), and a “rod” made of 62 alpha helices arranged into 13 domains (R1-R13) + a homodimerization domain (DD) that corresponds to the 62th alpha helix at the C-terminal end of the molecule . Here it is:
Fig. 8: Domain structure of talin. A, schematic diagram of talin showing the N-terminal FERM domain (F0, F1, F2, and F3 domains) linked via an unstructured region to the 13 amphipathic helical bundles of the talin rod, which terminates in a single helix, the dimerization domain (DD). Residue numbers for each domain (R1–R13) are shown. Helices are numbered and vinculin-binding sites are colored red. Domains corresponding to the new structures reported here are shaded. B–E, NMR structures of the talin R3, R4, R5, and R6 rod domains. Ribbon diagrams of representative low-energy structures show the overall topology of each bundle. F, model of talin showing the structures of all 18 domains. From: RIAM and Vinculin Binding to Talin Are Mutually Exclusive and Regulate Adhesion Assembly and Turnover , licensed under Creative Commons Attribution Non-Commercial License
Essentially, talin is the main protein that connects the intracellular tail of the beta subunit of integrin to actin. To do that, it has two integrin binding sites (one in F3 and 1 in R11) and three actin binding sites (one at F2-F3, one at R4-R8 and one at R13-DD). It has also domains for vinculin, its main co-interactor, and for many other proteins that are part of the Adhesome.
The working of talin is, of course, not completely understood, but much is known. I will try to sum up the essentials:
a) Talin has an important role in activating integrin and therefore the whole FA. It is therefore one of the main actors in the inside-out activation signaling shown at Fig. 3.
b) It is one of the most important protiens that can sense mechanical forces. IOWs, the application of mechanical forces can change its configuration and its biochemical affinities. In that sense, it works as a “hub” that integrates the many signals involved in the function of FAs, so much so that some have even introduced the concept of a “talin code”.
c) In the FA activation phase, talin ismore or less parallel to the cell membrane, linking up to 4 different integrin tails by its 4 integrin binding domains (in the homodimer), contributing to the clustering of integrins and to the growth of the FA.
d) In that same phase, it binds many actin molecules, that can provide a mechanical pull on the actin molecule both in a direction parallel to the call membrane and in a perpendicular direction. Those forces contribute to change talin conformation, and to its ability to recruit new proteins in the Adhesome, such as vinculin, RIAM, FAK, Paxillin and others.
e) In the mature FA, talin spans all three functional layers of the Adhesome, with its head near the cell membrane and the rod end in the actin regulatory layer.
Many of those features are described in detail in this recent paper:
Talin – the master of integrin adhesions
However, the most important point for our discussion is the second one: the ability of talin to repsond to the application of mechanical forces. We will go into greater detail about that in the section about mechanosensing.
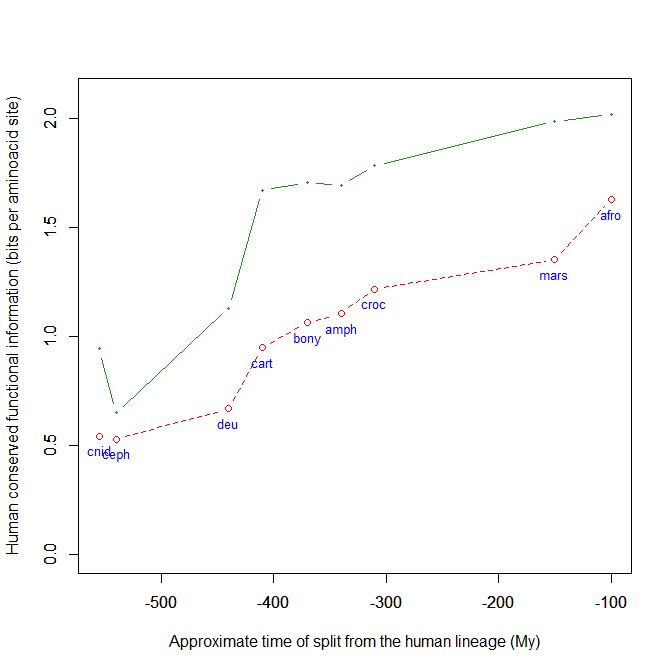
Finally, it can be interesting to mention that talin is an extremelly conserved protein. It shows almost 1 bit per AA conservation with human sequence already in cnidaria, and just a little bit more in pre-vertebrates. As the protein is so long, that amounts to about 2500 conserved bits. In vertebrates (cartilaginous fishes) it exhibits a well defined information jump (0.54 baa, corresponding to 1376 bits). That brings the molecule to an astonishing level of homology between cartilaginous fish and humans: 1.67 baa, 4250 bits. IOWs, the human protein and the protein in callorhincus milii (the ghost shark) share 2109 AA identities (83%) after 400+ million years of evolutionary separation!
Fig. 9: The evolutionary history of Talin shows extreme conservation in Metazoa and a definite information jump in vertebrates.
-
Mechanosensing
So, what is the purpose of all these complex structures, of all these intricate connections and potential regulations?
There are many answers to that, but the first and certainly one of the most important is: to perceive.
For the cell, its outer environment is everything: it is the reality where the destiny of each individual cell has to be implemented. But, as said, that outer environment is not a hostile and unpredictable world: it is a world made by the cells themselves, the Extracellular matrix, including of course the other cells in it.
Indeed, we have to clarify here that “the other cells” are an important part of the whole system. What we have said up to now is about the connection between each individual cell and the ECM. But there two more big and important subjects, and they are the connections, and the communications, between different cells.
Because adhesions are not realized only between a cell and the ECM. They exist between cells, too. And cell-cell adhesions are implemented by specific structures and proteins that are at least as complex as those that implement the cell-ECM adhesions. And yet very different from them.
In particular, cell-cell adhesions are not implemented by Integrins, but by completely different classes of proteins, such as the Cadherins. And of course there is a complex multi-protein structure associated to cadherins too, and it is called the Cadherin Adhesome. And there are different types of cell-cell juntions. And, as we can imagine at this point, cell-cell ahdesions do interact in many ways with the cell-ECM adhesions that we have been describing here.
However, to discuss these other aspects would certainly make this OP infinitely long, and so the discussion here will be “limited” to the cell-ECM interactions.
So, what does the cell need to know about its surrounding ECM?
It needs a lot of different things, certainly. Because, as we have seen, the ECM can be completely different in different parts of the organism. It has different biochemical composition and different physical properties. And both things are very important to the cell.
In a sense, biochemical composition is something that we can easily imagine as a target for the cell’s informational resources. We are accustomed to think that the cell works through membrane receptors, biochemical signaling pathways, and so on. And we have seen that at least part of the mechanisms we have described here are apparently “normal” biochemical affinities between a receptor and a ligand: in particual, the extracellular part of the integrin molecule certainly recognizes different ligands in the different proteins that are present in the ECM, by the active site formed by the two “heads”.
But what about mechanical information? We have seen that the binding to ligand has really the puprose to generate an adhesion to the ECM, which then matures into a very complex structure, the Focal Adhesion. It is not in itself a signal to the cell, but rather a tool to generate a signal. Because the signal that is to be generated is mechanical, not biochemical.
IOWs, the cell has to “perceive” the mechanical properties of the ECM, and also any specific mechanical forces that are applied to the ECM.
The complex mechanisms that implement that “perception” are called, globally, mechanosensing.
Now, here is where things become really complex, in the sense that we still don’t understand them. Everything about mechanosensing is, at present, poorly understood, and remains rather obscure. I will try to list just a few interesting points that are of paramount importance.
First of all, what are the physical properties of the ECM that are perceived by the cell? There are many of them, and all of them seem to be able to influence the cell and its reactions.
The best understood are:
- Rigidity or stiffness: the extent to which ECM resists deformation in response to an applied force.
- Elasticity: the ability of ECM to return to its original size and shape when a force is removed.
- Viscoelasticity: the association of viscous and elastic properties (gradual return to the original size and shape in time).
- Dimensionality: cells seem to be able to react in specific ways to 1D, 2D and 3D information.
- Strain-stiffening: ECM seems to be able to stiffen when forces are applied, either from the cell itself or from the outer environment.
- External forces applied to the ECM and transmitted to the cell, such as tension, compression, shear, swelling.
There seems to be a constant cross-talk between the cell and those mechanical forces: Focal Adhesions are constantly formed and dissolved in response to forces, and at the same time they transmit forces from the cell to the ECM, and the mechanical reaction of the ECM to those cell-generated forces is probably one of the main “messages” that the cell can translate into novel action.
The role of actin
Actin filaments have a very important role in all this, but again it is a role that is still poorly understood.
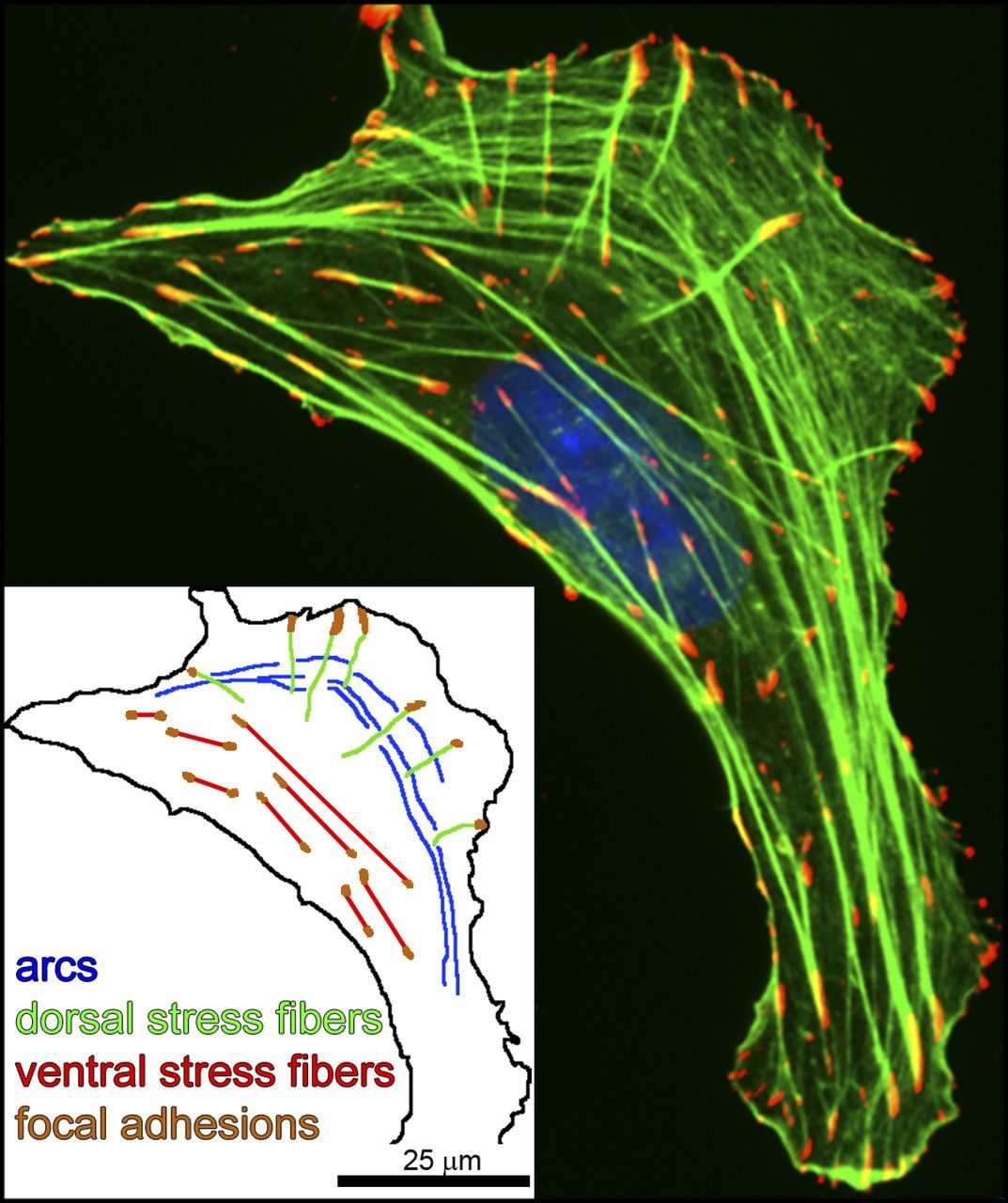
Actin polymerizes and forms specific structures. Of particular interest for our discussion are the “stress fibers”, long filaments of actin associated to other proteins, including myosin and alpha-actinin. Stress fibers can contract, and there are different types of them. The most relevant here are probably the ventral stress fibers, that are anchored to a Focal Adhesion at both ends.
Fig. 10: Three types of actin SFs. U2OS human osteosarcoma cells were plated on 10 µg/ml fibronectin-coated coverslips and allowed to attach and spread for 4 h before fixation (Hotulainen and Lappalainen, 2006). In the immunofluorescence image, antiphosphotyrosine was used as a marker for focal adhesions (red), phalloidin was used for F-actin SFs (green), and the nucleus (blue) was detected by DAPI. This single cell exhibits the three main types of actin SFs: (transverse) arcs, dorsal SFs, and ventral SFs. (inset) Schematic drawing depicting the SF subtypes. From: The tension mounts: Stress fibers as force-generating mechanotransducers , licensed under a Creative Commons License (Attribution–Noncommercial–Share Alike 3.0 Unported license, as described at http://creativecommons.org/licenses/by-nc-sa/3.0/
Moreover, when the cell moves actin is constantly polymerized at the leading edge of the cell, and moves towards the cell body, to be degraded at the trailing edge. That is the cause of the so called “actin retrograde flow”, which is not in itself a contractile movement, but is involved in both the formation of FAs and the cell migration, but in a complex, not well understood and somewhat paradoxical way (the direction of the actin flow is indeed opposite to the direction of the cell movement). Here is an amazing video of the retrograde flow:
-
Mechanotransduction
OK, so now we know that the cell has many complex ways to become aware of mechanical forces, generate them, perceive them, react to them.
But the problem remains: how does this mechanosensing relate to the more traditional world of cell functions, IOWs the biochemical world? How is mechanical information translated into biochemical sigmals, so that the two layers can communicate and interact?
The answer ot that is mechanotransduction: the translation of mechanical stimuli into biochemical signaling.
We have alredy seen that many proteins involved in the formation of FAs are capable of mechanosensing, first among them talin.
Let’s see more in detail one of the many ways that such mechanosensing can take place. Let’s go back to talin. The different bundles that make the rod of the molecule have different mechanosensitivity. One of the most mechanosensitive bundles is R3. In this domain, the folding forces are weaker, and they can be antagonized by the application of a mechanical force, which acts to unfold the sequence. That’s what happens when actin binds one of the actin binding sited of talin: the force generated by that binding “stretches” the talin molecule, and the R3 domain is the first to unfold. That partial unfolding “exposes” in R3 specific vinculin binding sites, which were previously hideen in the folded state. So, vinculin binds to talin, and the whole system progresses toward the formation of the FA.
This is just an example of how mechanical forces can generate biochemical activities, when the molecule is programmed in an appropriate way. But still, we are not seeing any connection to cell signaling.
The modifications in the cell that are initiated by mechanical signals are called, as a whole, mechanoresponse. They include a lot of different events, but of course most of the response has to be mediated by the fundamental hub of cell states: transcription regulation. Therefore, mechanical stimuli and their inherent information must, in some way, reach the nucleus.
There are two different modalities for that transmission:
Biochemical signaling.
This is the slower way, but it is extremely complex and can reach practically all aspects of cell activity. Indeed, the system of Integrins and Focal Adhesions has direct effects on many cell signaling pathways.
Many of those connections are mediated by phosphorylation networks, involving for example the Src family kinases, and the specific Adhesome protein FAK (Fochal Adhesion Kinase), plus many small GTPases, including RhoA.
The main signaling pathways that are modulated by mechanical signals deriving from FAs are:
- Hippo signaling, mainly mediated by the Yap-Taz proteins.
- Serum response factor (SRF) pathway.
- Wnt Receptor Signaling
- NF-kB pathway, directly activated by FAK
All these pathways are extremely complex, and they are at the very center of most cell activities, modulating cell transcription at all levels. All of them are strictly connected to mechanosensing, in many different ways that have been studied in great detail, but that are still poorly understood.
Mechanical signaling.
But there is a quicker way that can transmit information from FAs to the nucleus: direct mechanical signaling, from the cytoskeleton to the nucleoskeleton.
To accomplish that, a specific structure acts as interface between the two skeletons. It’s called LINC (linker of the nucleoskeleton and cytoskeleton) complex, and it includes the nuclear transmembrane protein emerin, the inner nuclear protein SUN and the nuclear lamina.
Again, in the nucleus, mechanical signals are translated into biochemical events, in particular deep chromatin rearrangements and epigenetic modifications, affecting all cell activities thorugh transcription regulation.
Some recent information about these complex topics can be found here:
Integrin-mediated mechanotransduction
In particular, the section: “Cellular responses in mechanotransduction”.
And a simpler summary can be found here:
Which biochemical pathways are regulated by mechanical signals?
Who makes the ECM?
And, of course, the cells themselves are the creators of the ECM. It’s the cells that synthesize the proteins, and all the components. The world where the cells reside and move is made by the cells themselves.
And, as we have seen, it is an extremely dynamic world. The structure and composition of the ECM is subject to constant changes and remodeling, and of course it’s the cells themselves that originate those changes. Different cells, in different ways. And the mechanoresponse in the cell to mechanical information from the ECM is certainly the guide to changes in ECM structure and composition that come from cell activity.
So, it’s a strict and constant crosstalk between the ECM and the cells that generate and remodel it. And hundreds of proteins, networks, components, structures, both in the cell and outside it, are involved in the process.
-
What about the cell?
So, we know that the cell uses mechanosensing and mechanotransdution to get information about the ECM and to connect to it. And then? What are the active responses of the cell?
As we have said, changes in the secretion of proteins and other components to change the composition and structure of the ECM are certainly an important aspect. But there is more.
As already anticipated, the interaction between the cell and the ECM has two very big purposes, one in stationary cells, and the other, the most important one, in cells that have to move.
Stationary cells
All cells are anchored in different ways to the ECM. That interaction has a definite and complex effect on cell morphology, and contributes to determine cell shape. Cell shape is also connected to cell fate, to mitosis and differentiation, even to apoptosis. All these processes are strongly conditioned by the interaction with the ECM by mechanosensing and mechanotransdution.
Here is a recent paper about these issues:
Mechanochemical Signaling Directs Cell-Shape Change
Abstract
For specialized cell function, as well as active cell behaviors such as division, migration, and tissue development, cells must undergo dynamic changes in shape. To complete these processes, cells integrate chemical and mechanical signals to direct force production. This mechanochemical integration allows for the rapid production and adaptation of leading-edge machinery in migrating cells, the invasion of one cell into another during cell–cell fusion, and the force-feedback loops that ensure robust cytokinesis. A quantitative understanding of cellmechanics coupled with protein dynamics has allowed us to account for furrow ingression during cytokinesis, a model cell-shape-change process. At the core of cell-shape changes is the ability of the cell‘s machinery to sense mechanical forces and tune the force-generating machinery as needed. Force-sensitive cytoskeletal proteins, including myosin II motors and actin cross-linkers such as α-actinin and filamin, accumulate in response to internally generated and externally imposed mechanical stresses, endowing the cell with the ability to discern and respond to mechanical cues. The physical theory behind how these proteins display mechanosensitive accumulation has allowed us to predict paralog-specific behaviors of different cross-linking proteins and identify a zone of optimal actin-binding affinity that allows for mechanical stress-induced protein accumulation. These molecular mechanisms coupled with the mechanical feedback systems ensure robust shape changes, but if they go awry, they are poised to promote disease states such as cancer cell metastasis and loss of tissue integrity.
Cell migration
However, the most complex procedure that is linked to the cell-ECM interaction is probably cell migration.
We all know that cells can move. Bacteria can move, single celled eukaryotes can move, and many different tools implement movement in those cells, including different kinds of flagella and amoeboid movement by pseudopodia. But cells in a multicellular organsism, as we have seen, exist and move in the ECM, and moving in the ECM is quite a specific task, very different from motion in the “exterior” world. That kind of movement is based on adhesions to the ECM, Fochal Adhesions and the role of cell cytoskeleton.
Again, we must acknowledge that movement of cells in the ECM is largely poorly understood, even with all the things we know today.
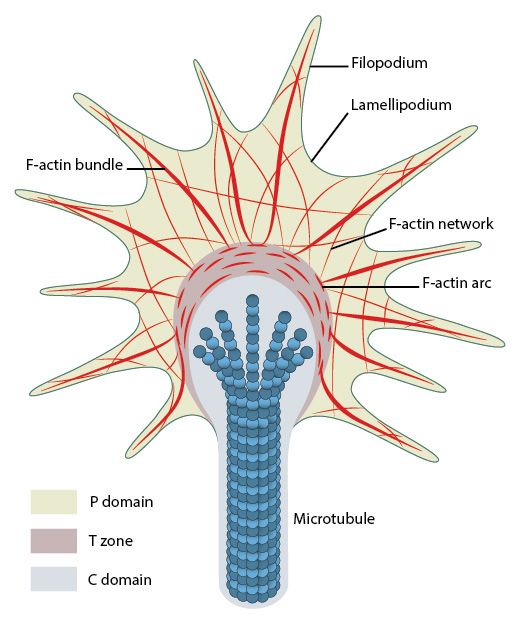
The main structure in the cell that implements movement is called lamellipodium. It is a flat, plate-like projecion at the leading edge of the cell, including an almost bidimensional actin network. Smaller spikes emerging from the cell edgle or from lamellipodia are called filopodia. Filopodia probably precede and guide the formation of lamellipodia. Focal Adhesions are usually rare or nascent at the tip of lamellipodia, while they are frequent and mature at the boundary between lamellipodium and cell body.
As said, the exact role of actin in generating the propulsion forces is not completely clear. I quote from this site:
Cell motility is driven by coordinated actin polymerization at the cell’s leading edge. However, it is still fiercely debated exactly how actin generates force to move a cell. The fine structure of filaments revealed by electron microscopy is exquisitely sensitive to the preparative methods used, and thus, various models have been proposed. Bringing together information from electron microscopy, live-cell imaging techniques, and super-resolution microscopy will be necessary to construct a definitive model.
Tha main idea is that actin generates the propulsion force in at least two different ways:
a) Contractile actin structures, like stress fibers, where actin is coupled to myosin and can therefore contract, should have a role. Different kinds of stress fibers have been implicated, but the general idea is that contraction is a component of the process.
b) The retrograde flow of actin, due to constant polymerization at the leading edge, would be one of the main sources of the forward propulsion, by some paradox effect linked to the resistance of Focal Adhesions at the rear of the lamellipodium. This is not a contractile mechanism, but it is connected to the polymerization of actin fibers at the leading edge, and its disassembling at the trailing edge. In this case, FAs would act as a “molecular clutch”, able to exist in a disengaged or engaged state, and therefore to connect the force to the propulsion in lamellipodia.
A good summary of these ideas can be found here:
How do focal adhesions act as molecular clutches in lamellipodia?
And this short video gives a good idea of the complex interactions between FA dynamics and the generation of lamellipodia for cell migration:
-
So, what is the purpose of all this functional complexity?
After all these complex details, let’s try to discuss the general scenario, and to draw some conclusions.
I think that there can be no doubt in anyone’s mind about the amazing complexity of the systems that have been described here. And remember, these are only a few aspects of what is known, and what is not known is certainly much more than what is known.
So the real question is: why? What is the purpose of all that?
To answer that, we must go back to our initial introduction. The real point is: everything depends on these processes, in multicellular organisms.
We know how complex and fascinating is the issue of cell differentiation, and how the many levels of control in transcription regulation certainly hold the key to that basic mystery.
But multicellular agents are not only the sum of differentiated cells. They are made by differentiated cell that build specific microscopic and macroscopic structures, in particular tissues and organs, in the context of the general body plan of the organism.
And remember, as all differentiated cell derive from the single cell that is the zygote, it is equally true that all body structures derive from that single cell, too.
So, when the original cell divides, at a certain point it generates spacial structures. Extracellular matrix comes into the game, hosting and guiding the cells. The basic 3 germ layers are produced, and then all the complex structures in the embryo, and then in the final body.
We don’t really know how all that is implemented, but we certainly know one thing: cells have to migrate to realize the plan. And they do that by moving in the ECM.
So, we can understand how the processes that regulate cell migration become the key to understand the development of a multicellular organism. That’s no small deal.
Cell migration
Indeed, cell migration is of paramount important in two different contexts:
a) Cells which have to migrate because of their intrinsic function. A good example of that are leucocytes and fibroblasts, involved in immune response, inflammation and wound healing. These cells migrate to reach the site where they have to accomplish their tasks.
b) Cell migration during embrional and fetal development. In this case, cells migrate to reach their final site in tissues and organs.
We will focus here on the second type of migration.
Cell migration starts with recognizing a direction. The cell must understand, in the 3D world of ECM, the correct direction it has to follow.
The direction generates three different important events:
- The establishment of the correct polarity: the cell defines a leading edge and a trailing edge, before strating to move, and as we have seen different structures are generated at the two poles.
- The initiation of propulsion in the right direction
- The maintenance and correction of propulsion throughout the pathway
Of course, the big question is: what guides the cells to their final abode? IOWs, what we observe is guided cell migration: cells do not wander randomly, but follow specific trajectories to reach specific places, in accord with their differention and their final roles. And, in doing that, they build highly ordered structures.
The good answer to that would be, again: we don’t really know in detail. However, a few things are known.
There are at least three types of factors influencing cell migration during development:
a) Chemotaxis
b) Haptotaxis
c) Mechanical properties of tyhe ECM (Mechanotaxis)
Electric forces can also have a role, but we will not discuss that aspect here.
Chemotaxis is the simplest concept. It means that some molecule dissolved in the extracellular fluid is recognized by the cell as attracting (or repulsing), and therefore the cell moves according to the concentration gradient of that substance. The important point here is that the chemotactic substance is in soluble form, and its gradient is a concentration gradient in the extracellular fluid, which, as we have said, is the fluid part of the ECM. Chemotaxis is probably more important in type a) migration, for example in inflammation, where inflammatory molecules attract leucocytes to the site of the lesion. In this case, cells are acctracted to a site, but there is no great specificity in that process.
Here is a paper about chemotaxis in organism development:
Chemokine-guided cell migration and motility in zebrafish development
Haptotaxis works in a similar way, but here the attractive (or repulsive) substrate is not soluble: it is, instead, bound to a surface, usually some of the ECM proteins, such as fibronectin and laminin. So, the cell interacts with the ligand through the complex Adhesion system that we have been describing.
Here is a paper about hapotaxis:
Haptotaxis is cell type specific and limited by substrate adhesiveness
Finally, it is well understood that the mechanical properties of the ECM, such as stiffness and so on, have a definite role in determining the path that the cell will follow (mechanotaxis), always by the mediation of the Integrin – FA system. The term “durotaxis”, for example, is used for the effects of stiffness.
Here is a recent paper about mechanotaxis:
Tissue mechanics regulate brain development, homeostasis and disease
The cell, in some way, integrates all those signals to decide what path it is going to follow.
Who sets the signals?
Of course, the cells themselves do that. Cells produce the ligands for both chemiotaxis and haptotaxis, and as we have seen it’s cells, again, that generate and remodel the ECM and determine its mechanical properties.
But the problem is: what cells?
In the case of chemotaxis, and probably haptotaxis, the simplest scenario is that the cells at the target site produce the ligands and, in some way, create the gradients. But, as we have seen, that is probably simpler with chemotaxis, because the ligand is soluble, while in haptotaxis a significant contribution of ECM proteins is more likely. Moreover, even intermediate cells (from sites between the migrating cell and the target site) can probably contribute.
How the mechanical properties of the ECM may be regulated in the pathway to be followed is less clear. It seems reasonable that many cell types hosted by the ECM itself could contribute to that, and possibly even the migrating cell itself.
As said, we do not understand enough yet. But one thing is certain: the migrating cell in some way can receive and integrate a lot of different signals, and in some way can control the complex activity that makes the propulsion possible (lamellipodia, actin activity, FAs and so on) to efficiently implement the correct migration.
A very good model: the axonal growth cone
A model that has been studied in great detail, and which shows many important aspects of the migration process, is the growth of the axon towards its destined synapsis, through the structure known as growth cone. Here, it’s not the whole cell that migrates, but rather its main extension, the axon, which is responsible for the main neuronal connection to another neuron. It is the main costituent of nerve fibers, and it is the structure used by neurons to output their signals to other cells.
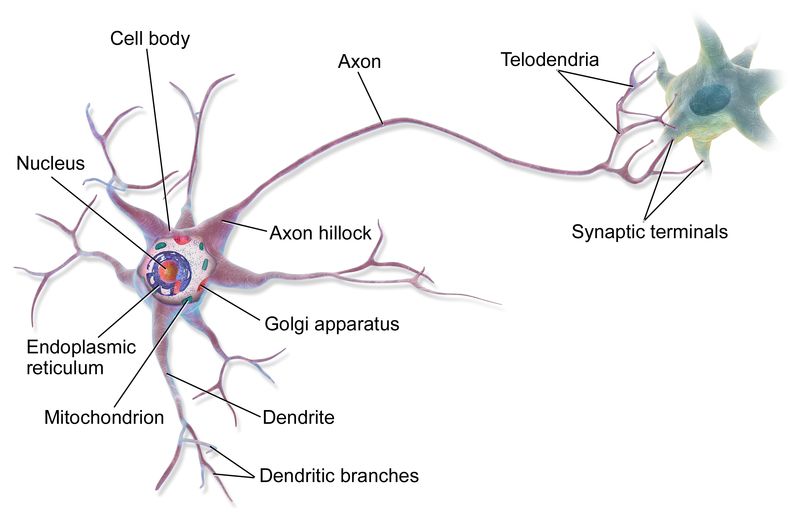
Fig. 11: A neuron and its axon.
Please, consider that axons can be very long: their length ranges, in humans, from less than 1 mm to more than 1 m. Each neuron has only one axon, that can innervate many target cells.
So, the axon grows from the neuronal body to its target synapse, and as we have said that pathway can be very long, even 1 meter. That’s a very long walk for a minuscule cell protrusion.
Moreover, axons must connect to the right synapses. Needless to say, the pattern of neuronal connections is what makes the nervous system the wonderful tool that it is, and not a random and useless mess. For example, moto neurons in the brain must connect with great precision to the corresponding second motor neurons in the spine, that connect to the appropriate muscle fibers.
So, how does the axon grow? Indeed, it is a good summary of what we have said before.
The structure that allows the growth is called growth cone. This structure was discovered by Cajal as early as 1890, and in the same year he declared that:
nerve fibers “adopt predetermined directions and establish connections with defined neural or extra neural elements… without deviations or errors, as if guided by an intelligent force”
Cajal, S. R. (1890). Notas anatómicas I. Sobre la aparición de las expansiones celulares en la médula embrionaria. Gac. Sanit. Barc. 12, 413–419.
Ah, the good old times of unbiased scientific research! 🙂
So, what is a growth cone? Let’s take a few concepts from this good internet page:
(The) growth cone (is) a specialized structure at the tip of the extending axon. Growth cones are highly motile structures that explore the extracellular environment, determine the direction of growth, and then guide the extension of the axon in that direction. The primary morphological characteristic of a growth cone is a sheetlike expansion of the growing axon at its tip called a lamellapodium. When examined in vitro, numerous fine processes called filopodia rapidly form and disappear from the terminal expansion, like fingers reaching out to touch or sense the environment (Figure 23.1). The cellular mechanisms that underlie these complex searching movements have become a focus of cell biological studies of axon growth and guidance. Such movements are thought to reflect rapid, controlled rearrangement of cytoskeletal elements—particularly molecules related to the actin cytoskeleton—which modulate the changes in growth cone shape and ultimately its course through the developing tissues.
So, it is a sheetlike structure including at least one lamellipodium and many filopodia, at the leading edge of axonal growth. Exactly as we have seen in cell migration.
While the axon cytoskeleton grows mainly by the elongation of microtubules, the growth cone is primarily an acti-myosin structure. Filopodia are continuously and dynamically generated and they test the surrounding microenvironment in search of attractive and repulsive cues. The final pathway followed by the axon is the result of the integration fo all those signals and of their many different effects on the various intracellular pathways discussed above.
All the mechanisms described in this OP can be seen at work: Focal Adhesions, retrograde actin flow, and so on.
Here is a schematic image of the growth cone from the appropriate MBINFO page:
Fig. 12; Organization of cytoskeletal components (actin filaments and microtubules) in the growth cone. From: What is axon guidance and the growth cone?, licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
And here is a rather detailed video about the axon cone:
Of course, all that is not simple. I really have to quote the following paper, even if it is not so recent (2012), because of its title:
Functional Complexity of the Axonal Growth Cone: A Proteomic Analysis
Functional complexity? Oh, yes!
Here is the abstract:
The growth cone, the tip of the emerging neurite, plays a crucial role in establishing the wiring of the developing nervous system. We performed an extensive proteomic analysis of axonal growth cones isolated from the brains of fetal Sprague-Dawley rats. Approximately 2000 proteins were identified at ≥99% confidence level. Using informatics, including functional annotation cluster and KEGG pathway analysis, we found great diversity of proteins involved in axonal pathfinding, cytoskeletal remodeling, vesicular traffic and carbohydrate metabolism, as expected. We also found a large and complex array of proteins involved in translation, protein folding, posttranslational processing, and proteasome/ubiquitination-dependent degradation. Immunofluorescence studies performed on hippocampal neurons in culture confirmed the presence in the axonal growth cone of proteins representative of these processes. These analyses also provide evidence for rough endoplasmic reticulum and reveal a reticular structure equipped with Golgi-like functions in the axonal growth cone. Furthermore, Western blot revealed the growth cone enrichment, relative to fetal brain homogenate, of some of the proteins involved in protein synthesis, folding and catabolism. Our study provides a resource for further research and amplifies the relatively recently developed concept that the axonal growth cone is equipped with proteins capable of performing a highly diverse range of functions.
OK, now let’s go to the proteins involved in the guidance of the axonal growth cone, because something is known about them. Here is an essential list:
- Ephrins and ephrin receptors. These are a well studied class of membrane bound ligands and recepors that have an important role in axon guidance mediated by cell-cell interaction. See here: Ephs and ephrins
- Netrins: a class of secreted chemotropic proteins that act as both attractants and repellents in axonal cone guidance.
- Semaphorins: a set of classes of proteins, both secreted and membrane bound, that usually act as repellents for axon growth.
- Plexins: the main receptors for semaphorins.
- Slits: ECM proteins secreted by structures in the nervous system, such as the floor plate or the midline glia, which act as midline repellents, preventing the crossing of axons through the midline of the central nervous system.
- Robo: the main receptors for Slits.
An interesting concept is that some axons act as pioneer axons: in a sense, they find a pathway and “open” it for other axons to follow.
And here is a recent (May 2018) review of those topics:
Understanding axon guidance: are we nearly there yet?
Let me quote a few titles from the paper sections:
- A richness of signals: redundancy of guidance information ensures correct navigation within the spinal cord
- Crosstalk between different families of guidance cues
- The regulation of axon guidance receptors at choice point
And finally I quote here the conclusions from the same paper (emphasis mine):
Perspectives: so, are we nearly there yet?
As I have highlighted here, our knowledge of neural circuit formation in the brain is still very much in its infancy. We can infer molecular mechanisms from what we have learned in one system to another but there is still not a single population of axons for which we have a complete understanding of the molecular mechanisms of navigation to the final target. So, we are clearly not there yet! A major challenge remains the characterization of the precise temporal regulation of guidance signals and the interactions between different signalling pathways that cooperate to guide axons to their intermediate, and ultimately final, targets. Axon guidance studies in a variety of organisms clearly indicate that the regulation of axon guidance signalling involves all possible mechanisms of regulation: transcriptional and translational control, trafficking of specific vesicles, and changes in protein-protein interactions as well as protein stability. Furthermore, the link between the interactions of guidance receptors and their ligands with the observed behaviour of growth cones is still missing. We also only have a very superficial understanding of the association between surface receptors and the regulation of cytoskeletal dynamics responsible for steering growth cones (Gomez and Letourneau, 2014). Similarly, our knowledge on specific intra-axonal trafficking of signals is poor. To make the next step in our understanding of axon guidance, it will be important to keep complexity in mind. Classical loss-of-function approaches might not reveal the complex interaction between guidance cues and their different receptors. Precise temporal control of experiments during embryonic development is difficult in mammals. Therefore, it will be important to make use of diverse animal models, each with its strengths and weaknesses. A particular challenge will be the visualization of the functional link between surface receptors and the cytoskeleton. This is becoming easier to do in vitro thanks to high-resolution imaging techniques, but in vitro experiments will not allow for the analysis of axon guidance as they will never be able to mimic the complexity of cell-cell interactions in the developing tissue. It is thus clear that understanding axon guidance remains a challenge, and that a multifaceted, multidisciplinary approach will be required to understand not only how a single axon finds its target but how billions of axons manage to do so.
Yes, billions. There are about 10^11 neurons in the human nervous system, and each of them has probably about 7000 connections.
-
Conclusions
So, what have we learned from this long discussion?
- The transition to metazoa implies a whole new world of functional complexity, a specific general plan that allows the implementation of tissues, organs, body plans.
- Part of it is, of course, cell differentiation: each cell must be guided to its final state, its final specific phenotype, starting from one common genome/epigenome. This was the subject of my previous OP about transcription regulation.
- But another important part of it is cell location, in time and space. The ordered guidance of each body cell to the right place and to the right functional connections is the foundation for the implementation of the general plan, which is made of specialized tissues that make specialized organs and consistent global organisms.
- Cells exist in a complex 3D environment created, maintained and dynamically restructured by the cells themselves: the ECM.
- The interaction between cells and the ECM is essential to many cell functions, starting with cell shape, and including cell migration in development and for other functional needs.
- The interaction between cells and the ECM is not only chemical, but essentially mechanic.
- The Integrins and the Focal Adhesions are an extremely dynamic system that links the ECM to the cytoskeleton, in particular to actin.
- Cell migration is realized by the integration of an extremely rich network of signals, both mechanical and biochemical. Cell-cell interactions have also a leading role.
- How that integration is achieved by each specific cell is still not understood.
- Indeed, a lot of the essential aspects of all these issues are still poorly understood.
- The axonal growth cone is another example of “migration” of a cell structure, which involves billions of specific pathways. The whole structure of the central nervous system depends on those processes.
- Whatever the mechanisms that we still don’t understand, it is rather clear that coordinated cell migration and axon growth require not only the amazing structures and integration abilities in each individual cell, but also the cooperation of many different actors: the migrating cell, the ECM, guiding proteins and ligands, intermediate cells, the final target, and probably many other components.
- To me, this is one of the most beautiful examples of coordinated, irreducible functional complexity: an implementation of design that can only inspire deep awe and wonder.