In Illustra Media’s Darwin’s Dilemma, there is a clip on proteins as islands of function in amino acid sequence space:
[youtube h38Xi-Jz9yk]
Food for thought.
As a stimulus to such, let us next note how the bloggist Wintery Knight has given an interesting summary of the challenges involved if a chance-dominated process is invoked for a hypothetical 100-AA polypeptide:
Let’s calculate the odds of building a protein composed of a functional chain of 100 amino acids, by chance. (Think of a meaningful English sentence built with 100 scrabble letters, held together with glue)
Sub-problems:
- BONDING: You need 99 peptide bonds between the 100 amino acids. The odds of getting a peptide bond is 50%. The probability of building a chain of one hundred amino acids in which all linkages involve peptide bonds is roughly (1/2)^99 or 1 chance in 10^30.
- CHIRALITY: You need 100 left-handed amino acids. The odds of getting a left-handed amino acid is 50%. The probability of attaining at random only L–amino acids in a hypothetical peptide chain one hundred amino acids long is (1/2)^100 or again roughly 1 chance in 10^30.
- SEQUENCE: You need to choose the correct amino acid for each of the 100 links. The odds of getting the right one are 1 in 20. Even if you allow for some variation, the odds of getting a functional sequence is (1/20)^100 or 1 in 10^65.
The final probability of getting a functional protein composed of 100 amino acids is 1 in 10^125. Even if you fill the universe with pre-biotic soup, and react amino acids at Planck time (very fast!) for 14 billion years, you are probably not going to get even 1 such protein. And you need at least 100 of them for minimal life functions, plus DNA and RNA.
But, some will object, it’s not just chance involved!
That is, they are appealing to self-organisation and/or mechanical necessity, or incremental complexification — a sort of pre-life evolution.
Especially, with RNA acting as a catalyst and potential information store.
Mechanical necessity, a forced sequence of bonds, is both counter to what we know of the chemistry of both AA and D/RNA chaining, and would undermine the flexibility of sequence required. Templating, in which a clay bed or the like would force the sequence only displaces the informational specificity one step back.
And while pebbles on Chesil Beach are sorted by size along the beach and while similar self-ordering phenomena for instance explain hurricanes and the hexagonal polar cloud ring on one of our gas giant planets, that is not even comparable to the functional and aperiodic, non sorted sequence chaining we need for the information-rich polymers of life.
Functional sequence specificity simply is not explained by forces of order or of randomness, once we have to deal with relevantly complex sequences and organised clusters of hundreds or thousands of molecules required to achieve cellular function.
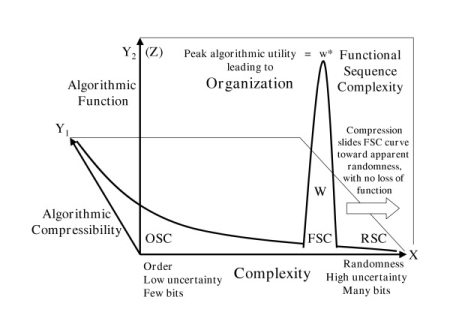
They don’t even plot to the same zones on a graph of random, ordered and functional sequence complexity, as Trevors and Abel remind us from 2005:
No wonder, the making of proteins in the cell is such a meticulous, organised step by step process:
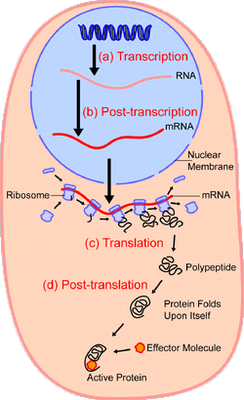
Zooming in on the ribosome:
Let us remind ourselves: cells are gated, encapsulated, metabolic automata with self maintenance, responsiveness to environment and a code based self replication. As Mignea summed up in the context of requisites for a minimally functional, self-replicating cell:
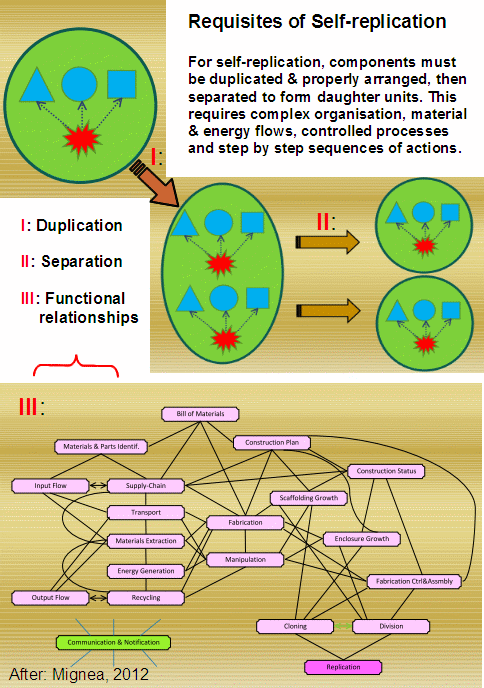
That means, there is a stiff irreducible complexity threshold involved, which locks out incrementalist solutions, as we confront Menuge’s criteria C1 – 5 to get incrementally additive cobbling together (NB: Menuge set these in the context of the flagellum, but the challenge is much broader in relevance as anyone struggling to get a rare car part can tell us):
For a working [bacterial] flagellum to be built by exaptation, the five following conditions would all have to be met:
C1: Availability. Among the parts available for recruitment to form the flagellum, there would need to be ones capable of performing the highly specialized tasks of paddle, rotor, and motor, even though all of these items serve some other function or no function.
C2: Synchronization. The availability of these parts would have to be synchronized so that at some point, either individually or in combination, they are all available at the same time.
C3: Localization. The selected parts must all be made available at the same ‘construction site,’ perhaps not simultaneously but certainly at the time they are needed.
C4: Coordination. The parts must be coordinated in just the right way: even if all of the parts of a flagellum are available at the right time, it is clear that the majority of ways of assembling them will be non-functional or irrelevant.
C5: Interface compatibility. The parts must be mutually compatible, that is, ‘well-matched’ and capable of properly ‘interacting’: even if a paddle, rotor, and motor are put together in the right order, they also need to interface correctly.
( Agents Under Fire: Materialism and the Rationality of Science, pgs. 104-105 (Rowman & Littlefield, 2004). HT: ENV.)
No wonder, then, that a few years back, Berlinski commented:
At the conclusion of a long essay, it is customary to summarize what has been learned. In the present case, I suspect it would be more prudent to recall how much has been assumed:
First, that the pre-biotic atmosphere was chemically reductive; second, that nature found a way to synthesize cytosine; third, that nature also found a way to synthesize ribose; fourth, that nature found the means to assemble nucleotides into polynucleotides; fifth, that nature discovered a self-replicating molecule; and sixth, that having done all that, nature promoted a self-replicating molecule into a full system of coded chemistry.
These assumptions are not only vexing but progressively so, ending in a serious impediment to thought. That, indeed, may be why a number of biologists have lately reported a weakening of their commitment to the RNA world altogether, and a desire to look elsewhere for an explanation of the emergence of life on earth. “It’s part of a quiet paradigm revolution going on in biology,” the biophysicist Harold Morowitz put it in an interview in New Scientist, “in which the radical randomness of Darwinism is being replaced by a much more scientific law-regulated emergence of life.”
Morowitz is not a man inclined to wait for the details to accumulate before reorganizing the vista of modern biology. In a series of articles, he has argued for a global vision based on the biochemistry of living systems rather than on their molecular biology or on Darwinian adaptations. His vision treats the living system as more fundamental than its particular species, claiming to represent the “universal and deterministic features of any system of chemical interactions based on a water-covered but rocky planet such as ours.”
This view of things – metabolism first, as it is often called – is not only intriguing in itself but is enhanced by a firm commitment to chemistry and to “the model for what science should be.” It has been argued with great vigor by Morowitz and others. It represents an alternative to the RNA world. It is a work in progress, and it may well be right. Nonetheless, it suffers from one outstanding defect. There is as yet no evidence that it is true . . .
Origin of life is the root of the tree of life, and so that is the decisive issue:
No roots, no tree.
And, on blind watchmaker mechanisms, there is no good reason to expect a root to form.
Where also, there is one reliably known adequate causal source for functionally specific complex organisation and/or associated information (FSCO/I).
Design.
Which is in the main being disregarded on a priori ideological grounds such as Lewontin so tellingly documented:
. . . It is not that the methods and institutions of science somehow compel us to accept a material explanation of the phenomenal world, but, on the contrary, that we are forced by our a priori adherence to material causes to create an apparatus of investigation and a set of concepts that produce material explanations, no matter how counter-intuitive, no matter how mystifying to the uninitiated . . . [[“Billions and billions of demons,” NYRB, Jan 1997. (If you imagine this is quote-mining, as a common how dare you cite that talking point alleges, cf. the fuller cite and notes here.)]
No wonder ID thinker Philip Johnson replied:
For scientific materialists the materialism comes first; the science comes thereafter. [[Emphasis original] We might more accurately term them “materialists employing science.” And if materialism is true, then some materialistic theory of evolution has to be true simply as a matter of logical deduction, regardless of the evidence. That theory will necessarily be at least roughly like neo-Darwinism, in that it will have to involve some combination of random changes and law-like processes capable of producing complicated organisms that (in Dawkins’ words) “give the appearance of having been designed for a purpose.”. . . . The debate about creation and evolution is not deadlocked . . . Biblical literalism is not the issue. The issue is whether materialism and rationality are the same thing. Darwinism is based on an a priori commitment to materialism, not on a philosophically neutral assessment of the evidence. Separate the philosophy from the science, and the proud tower collapses. [[Emphasis added.] [[The Unraveling of Scientific Materialism, First Things, 77 (Nov. 1997), pp. 22 – 25.]
There is no good reason why design is not sitting at the table as a serious explanatory candidate for the FSCO/I in first cell based life, and therefore also, in explaining the origin of major body plans. Motive mongering games and dismissive remarks notwithstanding.
In short, it is time for a serious re-think on the science of origins. END