Dr E. Selensky occasionally requests that UD posts an article on his behalf. What follows is his latest:
______________
Arguably, the RNA world model is excessively complex: it operates too complex structures and involves too complex interactions.
The origin of life, some researchers believe, must have been simpler.
In an attempt to close the gap between chemistry and life by naturalistic means a new model has been proposed recently, yet another one of many, that seeks to explain the rise of RNAs. This model is called template-assisted ligation. It has been proposed by Alexey Tkachenko and Sergei Maslov at American Institute of Physics. They hope it can help shed light on what could have preceded the RNA world.
The crux of the model is as follows. In an environment with cyclic parameter variations (the authors suggest that day-night variations of pH, salinity, luminosity etc. might have occurred in the hypothetical primordial ocean) small-sized polymers could form larger complementary polymer strands that then would undergo cyclic dissociations and associations. The transition from small-sized molecules to larger polymers is hysteretic and, once it happened under suitable conditions, it would no longer be required to maintain the replication of complementary polymer strands.
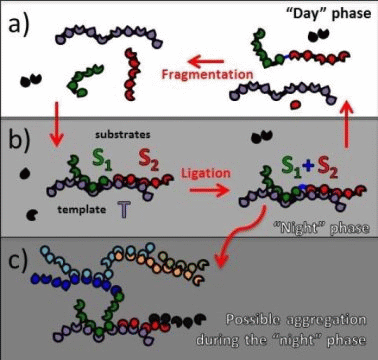
With all due respect, the model falls short of explaining the information processing capability of the living systems.
Autocatalytic replication by itself could not have given rise to a translation from the language of nucleotides to the language of peptides, which takes place in all known organisms today. Template-based replication is observed even in crystals and does not require translation in the above sense. On the contrary, such translation assumes the existence of logical relationships between the code and its referent (see Genetic code). These relationships are categorically different from physico-chemical regularities, which act as the key driver in the appearance of chemical replication in the model under consideration. The realization of the said logical relationships is only possible where the structure of the polymer, recognized by the translator as code, is inert or degenerate towards, arbitrary in relation to and does not chemically depend on the system dynamics.
The genetic information translation system that can be universally observed today includes three major physico-chemically independent components:
-
The code (nucleic acids);
-
The protocol (aminoacyl tRNA synthetases with their tRNAs);
-
The referents (polypeptides).
Additionally, the genetic information translation system is characterized by what is known as semantic closure (semantic self-sufficiency) whereby the code, apart from the other components of the next generation organism, also specifies the translation system itself that will have to recognize and interpret this same code in the subsequent generations. As far as I could see, the template-assisted ligation model does not even address the problem of the origin of biological information translation. It merely illustrates a mechanism of autocatalytic replication.
Many, if not all, known abiogenetic models (chemotons, hyper-cycles, RNA-world, RNA-peptide world, metabolic pathways or template-assisted ligation) fail to recognize that in reality there is no unintelligent non-telic physico-chemical pathway from organic chemistry to life. Information translation — a key property of living systems — is not a consequence of physical or chemical regularities but a result of the organization of the system, in other words, the boundary conditions on system dynamics.
The notoriously evasive complexity leap from organic chemistry to life defies attempts to ‘spread it out’ over a succession of elementary chemical interactions. Simple models do not take into account the necessary starting complexity of life as a whole, while more complex models depart from the original idea of non-telic non-agent-based abiogenetic synthesis: the more complex the model, the further it defeats the naturalistic purpose to represent life as ‘just a bit more complicated’ chemistry.
Finally, let me suggest some further reading.
-
David Abel: Examining specific life-origin models for plausibility, Academia.edu.
-
Edward Clark, Simon J. Hickinbotham, Susan Stepney: Semantic closure demonstrated by the evolution of a universal constructor architecture in an artificial chemistry. Published 17 May 2017, DOI: 10.1098/rsif.2016.1033.
And, a couple of my previous OPs:
_______________
Serious food for thought on OoL. END