Let us use a handy diagram of protein synthesis:
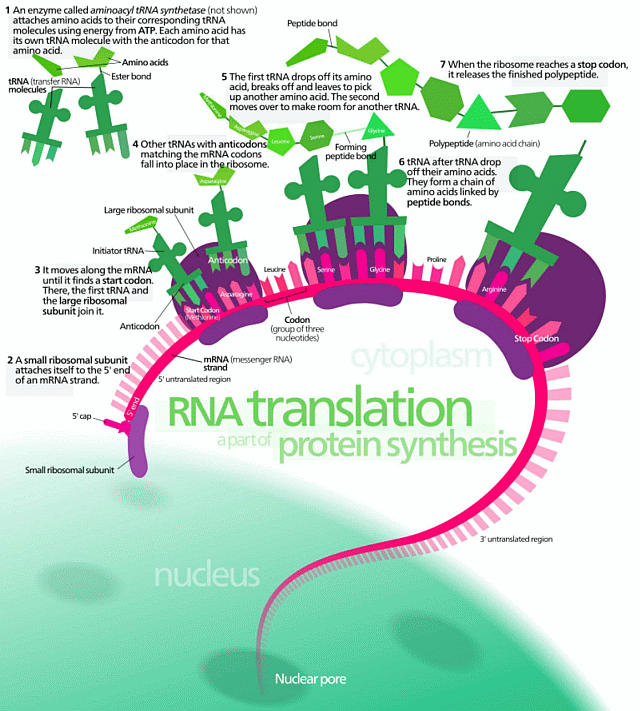
[U/D, Sep 2:] Where, to clarify key terms, let us note a key, classic text, Lehninger, 8th edn:
“The information in DNA is encoded in its linear (one-dimensional) sequence of deoxyribonucleotide subunits . . . . A linear sequence of deoxyribonucleotides in DNA codes (through an intermediary, RNA) for the production of a protein with a corresponding linear sequence of amino acids . . . Although the final shape of the folded protein is dictated by its amino acid sequence, the folding of many proteins is aided by “molecular chaperones” . . . The precise three-dimensional structure, or native conformation, of the protein is crucial to its function.” [Principles of Biochemistry, 8th Edn, 2021, pp 194 – 5. Now authored by Nelson, Cox et al, Lehninger having passed on in 1986. Attempts to rhetorically pretend on claimed superior knowledge of Biochemistry, that D/RNA does not contain coded information expressing algorithms using string data structures, collapse. We now have to address the implications of language, goal directed stepwise processes and underlying sophisticated polymer chemistry and molecular nanotech in the heart of cellular metabolism and replication.]
Let’s add more from Lehninger and heirs, who went out of their way to make the point, Oct 29:

Further u/d Sept 2: A look at creating mRNA:
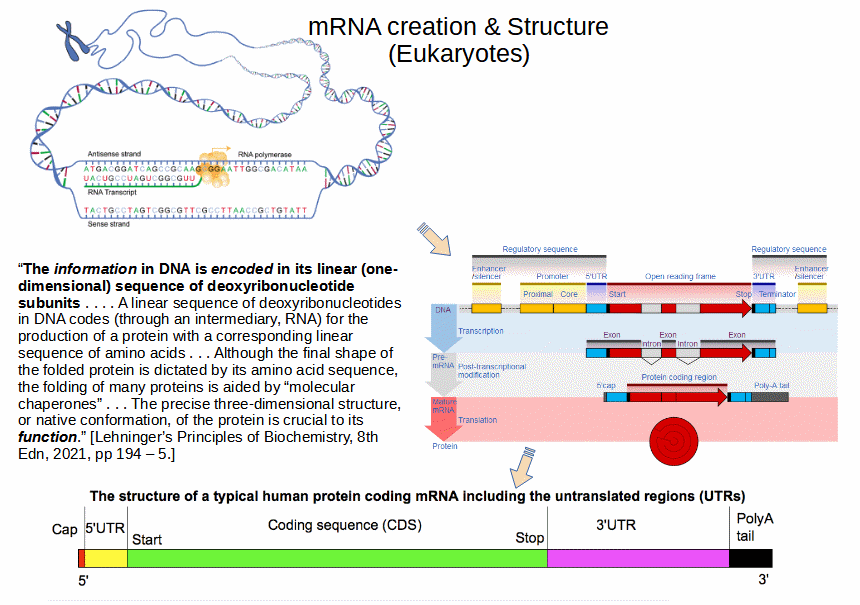
As Wikipedia admits (but, revealingly, does not duly emphasise) with a few telling words:
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription [–> algorithmic start, notice, transcribing is generally stepwise], it also guides the nucleotides into position, facilitates attachment and elongation [–> notice direct parallel to stepwise synthesis of AA chains for proteins, cf Lehninger 8th edn p 3346: “RNA polymerase elongates an RNA strand by adding ribonucleotide units to the 3 ′- hydroxyl end, building RNA in the 5 ′ → 3 ′ direction”], has intrinsic proofreading and replacement capabilities [–> language], and termination recognition [–> algorithmic halting] capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides.
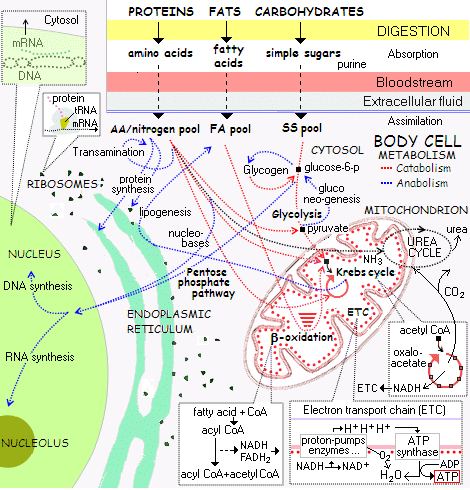
This is a corner of the general cell metabolism framework:
Where, Yockey observes (highlighted and annotated):
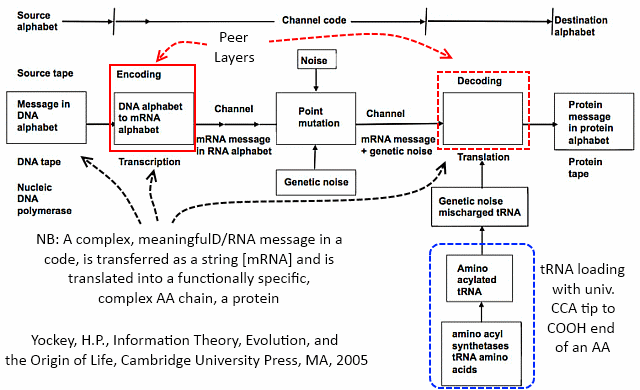
Where, too, the genetic code is, and its context of application is:
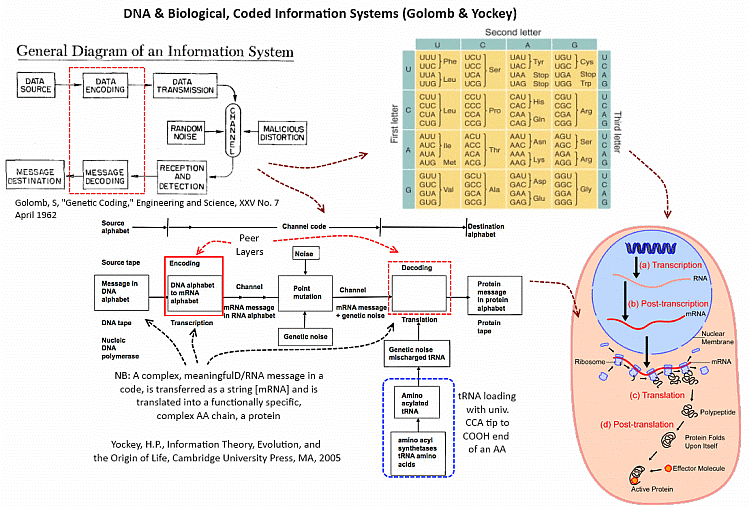
Given, say, Crick:

. . . we need to ask, why. END
F/N1: It seems advisable to highlight the layer cake architecture of communication systems
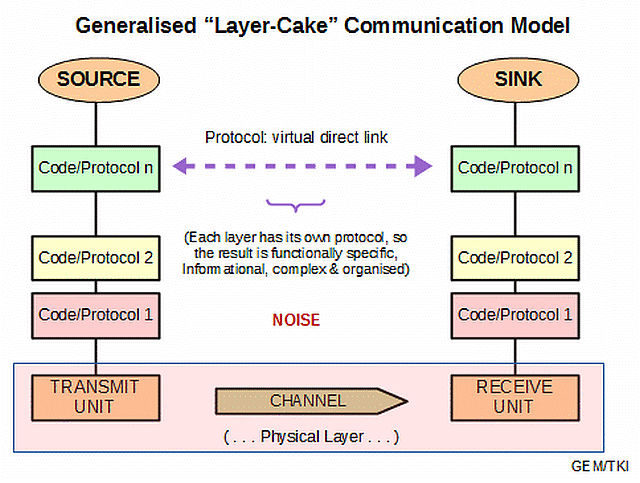
, , , and of Computers, following Tanenbaum:
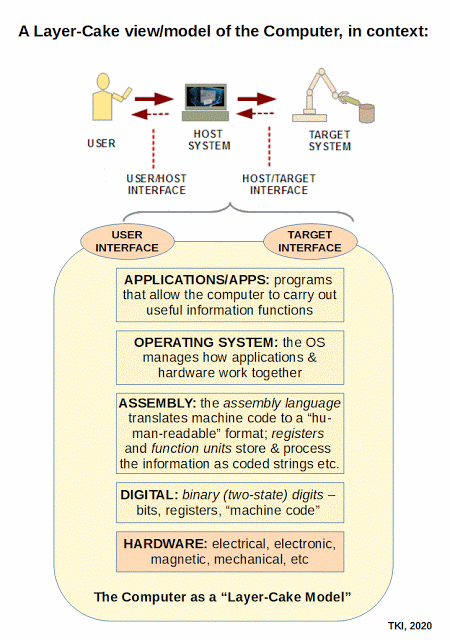
Clearly, the communication framework does not reduce to the physics of the hardware involved.
F/N2: Dawkins admits
He tries to deflect the force by appeal to “natural selection,” but protein synthesis and linked metabolic processes are causally antecedent to self replication and therefore pose a chicken before egg challenge especially for origin of life.