From ScienceDaily:
The world of epigenetics — where molecular ‘switches’ attached to DNA turn genes on and off — has just got bigger with the discovery by a team of scientists from the University of Cambridge of a new type of epigenetic modification.
…
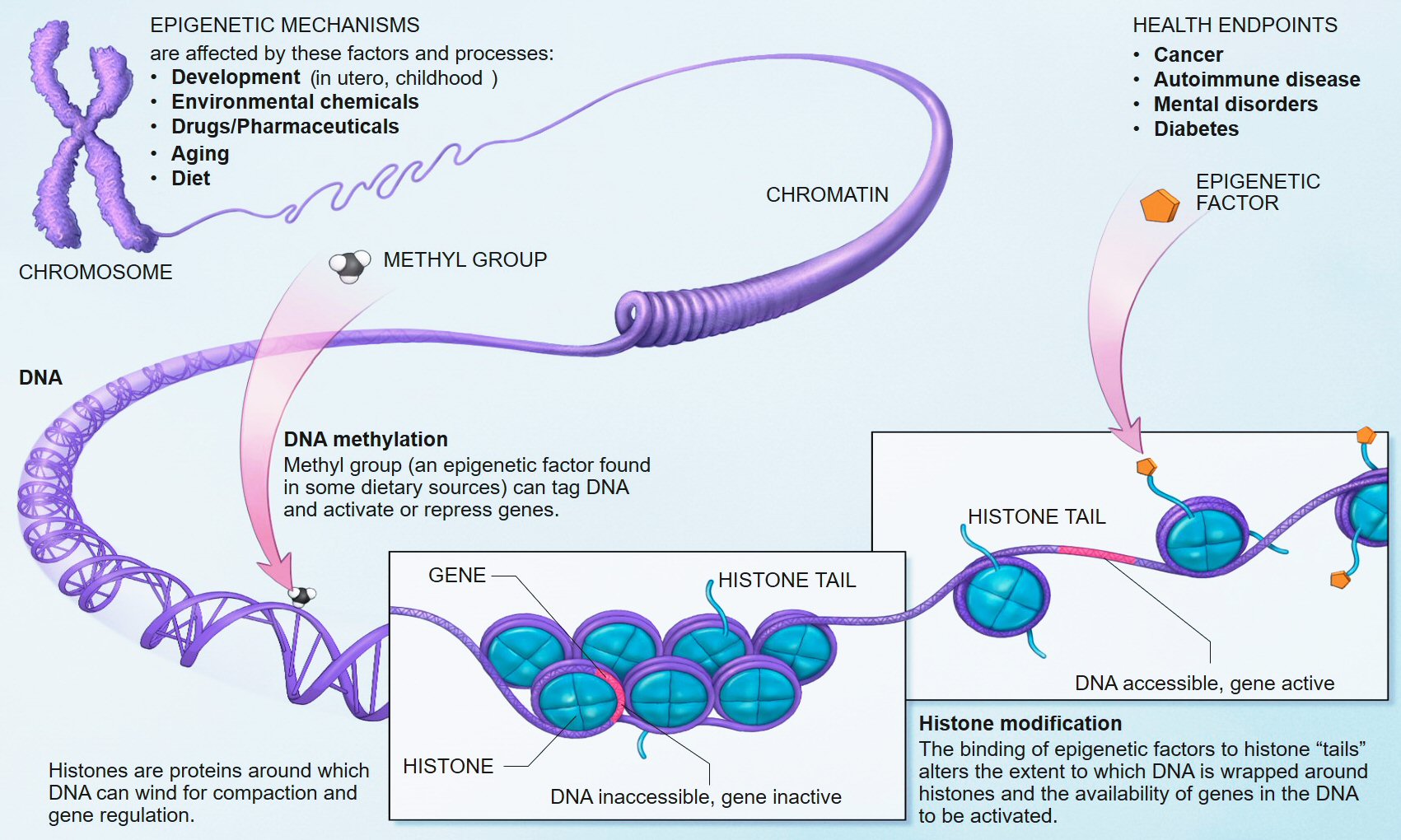
Epigenetics has so far focused mainly on studying proteins called histones that bind to DNA. Such histones can be modified, which can result in genes being switched on or of. In addition to histone modifications, genes are also known to be regulated by a form of epigenetic modification that directly affects one base of the DNA, namely the base C. More than 60 years ago, scientists discovered that C can be modified directly through a process known as methylation, whereby small molecules of carbon and hydrogen attach to this base and act like switches to turn genes on and off, or to ‘dim’ their activity. Around 75 million (one in ten) of the Cs in the human genome are methylated.
Now, researchers at the Wellcome Trust-Cancer Research UK Gurdon Institute and the Medical Research Council Cancer Unit at the University of Cambridge have identified and characterised a new form of direct modification — methylation of the base A — in several species, including frogs, mouse and humans.
…
“It’s possible that we struck lucky with this modifier,” says Dr Koziol, “but we believe it is more likely that there are many more modifications that directly regulate our DNA. This could open up the field of epigenetics.” More.
See also: Epigenetic change: Lamarck, wake up, you’re wanted
in the conference room!
Follow UD News at Twitter!
Note: Add “more diverse than thought” category to the list that includes “more complex than thought,” and “earlier than thought”
Here’s the abstract:
Methylation of cytosine deoxynucleotides generates 5-methylcytosine (m5dC), a well-established epigenetic mark. However, in higher eukaryotes much less is known about modifications affecting other deoxynucleotides. Here, we report the detection of N6-methyldeoxyadenosine (m6dA) in vertebrate DNA, specifically in Xenopus laevis but also in other species including mouse and human. Our methylome analysis reveals that m6dA is widely distributed across the eukaryotic genome and is present in different cell types but is commonly depleted from gene exons. Thus, direct DNA modifications might be more widespread than previously thought. (paywall) – Magdalena J Koziol, Charles R Bradshaw, George E Allen, Ana S H Costa, Christian Frezza, John B Gurdon. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nature Structural & Molecular Biology, 2015; DOI: 10.1038/nsmb.3145