One of the more peculiar objections to the design inference is the strident, often repeated claim that the genetic code is not a code, and that DNA and mRNA are not storing algorithmic, coded information used in protein synthesis. These are tied to the string (yes, s-t-r-i-n-g) data structure, a key foundational array for information storage, transfer and application. So, it seems useful to address the string as a key first principles issue, with the onward point being that strings of course can and do store coded information.
Let us begin with, what a string — yes, s-t-r-i-n-g — is (though that should already be obvious from even the headline):
Geeks for Geeks: A string is a sequence of characters, often used to represent text. In programming, strings are a common data type and are used for a variety of tasks, such as representing names, addresses, and other types of information.
Wikipedia confesses: In computer programming, a string is traditionally a sequence of characters, either as a literal constant or as some kind of variable. The latter may allow its elements to be mutated and the length changed, or it may be fixed (after creation). A string is generally considered as a data type and is often implemented as an array data structure of bytes (or words) that stores a sequence of elements, typically characters, using some character encoding. String may also denote more general arrays or other sequence (or list) data types and structures . . . . A primary purpose of strings is to store human-readable text, like words and sentences. Strings are used to communicate information from a computer program to the user of the program. A program may also accept string input from its user. Further, strings may store data expressed as characters yet not intended for human reading . . . . Example strings and their purposes . . . Alphabetical data, like “
AGATGCCGT” representing nucleic acid sequences of DNA . . .
So, it should not be surprising to see that DNA and RNA can store strings of information-bearing elements:

Where, of course, the genetic code is expressed in such strings. The (standard) code, mRNA form is:
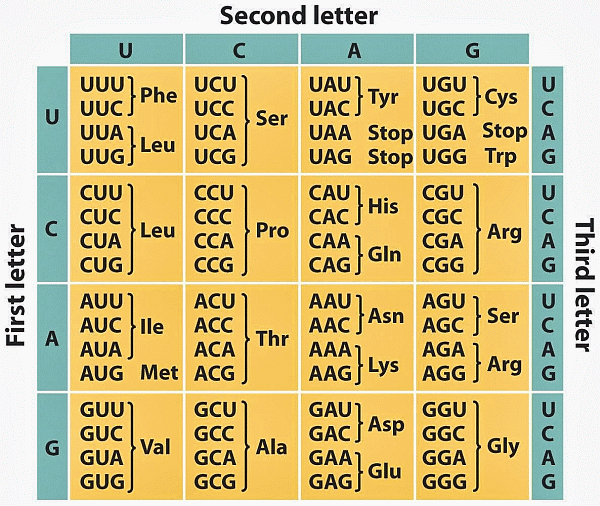
For very simple example, HT Khan Academy:

Of course, the above is the mRNA form, which would be transcribed and edited to cut out introns, and it leaves out onward complexities. For example, we can see how Insulin has two strands of AA’s interconnected through di-sulphide bonds, making up a 51 AA protein:

The end-product insulin protein is put together from the preproinsulin produced stepwise in the ribosome, by way of a clever alignment that uses a third, “scaffolding,” chain C sequence:

Using the code one could in principle back-translate to mRNA, however, in the DNA there are intervening Introns between the Exons expressed in the ribosome, so the human genome sequence is:

So, as usual, we see how sophisticated life is at molecular level. That said, we also see that as a key stage of protein synthesis, as ribosomes, mRNA and tRNA interact (with a complex cast of supporting molecules) AA chains are assembled with start, elongate, stop, executing a code driven algorithm. Where, AmHD defines:
[Algorithm:] A finite set of unambiguous instructions that, given some set of initial conditions, can be performed in a prescribed sequence to achieve a certain goal and that has a recognizable set of end conditions.
Illustrating:
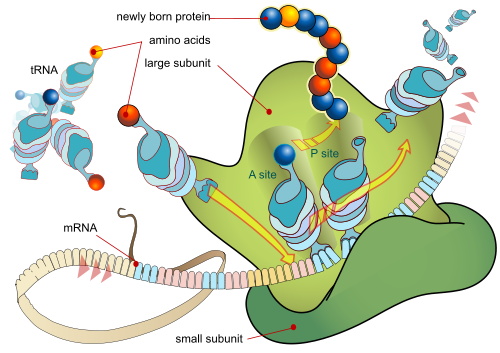
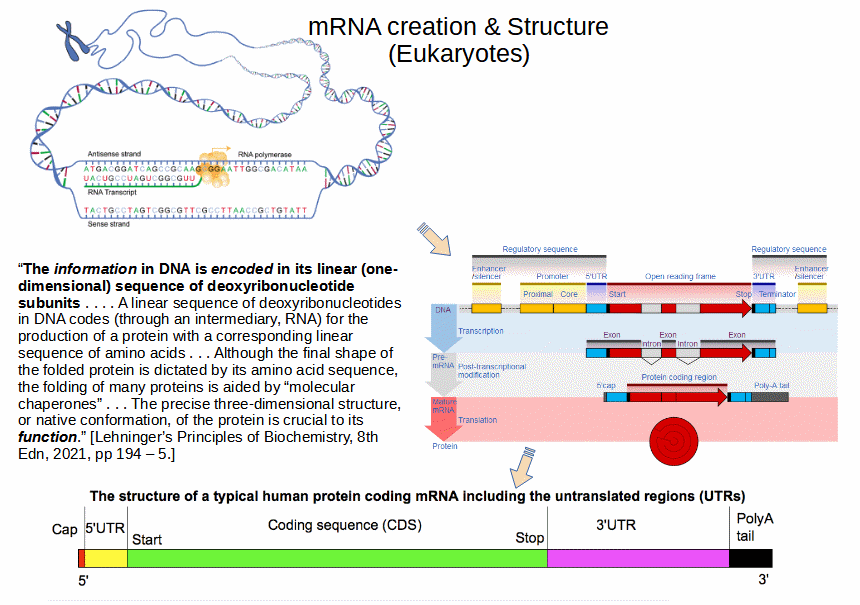
That should be enough to show the unbiased mind that coded algorithms are in the cell, and that DNA and mRNA act as string data structures. However, there are those who have proved resistant to such commonplace summaries or to citations from the sort of panels of experts who write major textbooks in biochemistry. For record, notwithstanding, here are Lehninger and heirs:

Lehninger and heirs go on to say, pp. 194 – 5:
We may also now observe a Nobel Prize Laureate, Sydney Brenner, in his article, Life’s code script . . . yes, it’s that obvious, published in 2012 in the leading Science Journal, Nature:
[Brenner:] ” . . . The most interesting connection with biology, in my view, is in Turing’s most important paper: ‘On computable numbers with an application to the Entscheidungsproblem’5, published in 1936, when Turing was just 24.
Computable numbers are defined as those whose decimals are calculable by finite means. [–> that is, effectively, by algorithms] Turing introduced what became known as the Turing machine to formalize the computation. The abstract machine is provided with a tape [–> with marks on it], which it scans one square at a time, and it can write, erase or omit symbols. The scanner may alter its mechanical state, and it can ‘remember’ previously read symbols. Essentially, the system is a set of instructions written on the tape, which describes the machine. Turing also defined a universal Turing machine, which can carry out any computation for which an instruction set can be written — this is the origin of the digital computer. [–> there is also, a more powerful oracle machine, capable of one step decisions]
Turing’s ideas were carried further in the 1940s by mathematician and engineer John von Neumann, who conceived of a ‘constructor’ machine capable of assembling another according to a description. A universal constructor with its own description would build a machine like itself. To complete the task, the universal constructor needs to copy its description and insert the copy into the offspring machine. Von Neumann noted that if the copying machine made errors, these ‘mutations’ would provide inheritable changes in the progeny.
Arguably the best examples of Turing’s and von Neumann’s machines are to be found in biology. Nowhere else are there such complicated systems, in which every organism contains an internal description of itself. The concept of the gene as a symbolic representation of the organism — a code script — is a fundamental feature of the living world and must form the kernel of biological theory. [–> note, again, author, context and publisher]
Turing died in 1954, one year after the discovery of the double-helical structure of DNA by James Watson and Francis Crick, but before biology’s subsequent revolution. Neither he nor von Neumann had any direct effect on molecular biology, but their work allows us to discipline our thoughts about machines, both natural and artificial.
Turing invented the stored-program computer, and von Neumann showed that the description is separate from the universal constructor. [–> that ‘description’ of course is encoded] This is not trivial. Physicist Erwin Schrödinger confused the program and the constructor in his 1944 book What is Life?, in which he saw chromosomes as “architect’s plan and builder’s craft in one”. This is wrong. The code script contains only a description of the executive function, not the function itself.
That’s why Yockey adapted Shannon’s architectural diagram for communication systems:
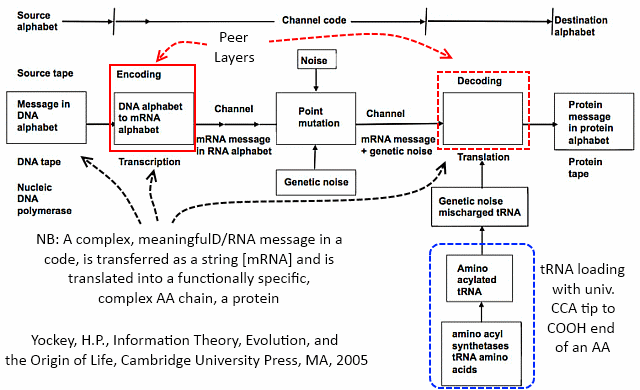
So, we may freely understand that DNA and associated molecules such as mRNA express string data structures, store coded biological information, that such information as used in protein synthesis expresses algorithms, and that therefore we are dealing with computation and associated computer language in the course of protein synthesis.
We may quote a Wiki confession:
[Wiki confesses:] Since 2001, 40 non-natural amino acids have been added into proteins by creating a unique codon (recoding) and a corresponding transfer-RNA:aminoacyl – tRNA-synthetase pair to encode it with diverse physicochemical and biological properties in order to be used as a tool to exploring protein structure and function or to create novel or enhanced proteins.[22][23]
H. Murakami and M. Sisido extended some codons to have four and five bases. Steven A. Benner [–>another guy] constructed a functional 65th (in vivo) codon.[24]
In 2015 N. Budisa, D. Söll and co-workers reported the full substitution of all 20,899 tryptophan residues (UGG codons) with unnatural thienopyrrole-alanine in the genetic code of the bacterium Escherichia coli.[25]
In 2016 the first stable semisynthetic organism was created. It was a (single cell) bacterium with two synthetic bases (called X and Y). The bases survived cell division.[26][27]
In 2017, researchers in South Korea reported that they had engineered a mouse with an extended genetic code that can produce proteins with unnatural amino acids.[28]
In May 2019, researchers reported the creation of a new “Syn61” strain of the bacterium Escherichia coli. This strain has a fully synthetic genome that is refactored (all overlaps expanded), recoded (removing the use of three out of 64 codons completely), and further modified to remove the now unnecessary tRNAs and release factors. It is fully viable and grows 1.6× slower than its wild-type counterpart “MDS42”
Indeed, the function of DNA as an information storage entity is so well established, that as Wiki also confesses, it has been adapted to general archival storage:
DNA digital data storage is the process of encoding and decoding binary data to and from synthesized strands of DNA.[1][2]
While DNA as a storage medium has enormous potential because of its high storage density, its practical use is currently severely limited because of its high cost and very slow read and write times.[3]
In June 2019, scientists reported that all 16 GB of text from Wikipedia’s English-language version had been encoded into synthetic DNA.[4] In 2021, scientists reported that a custom DNA data writer had been developed that was capable of writing data into DNA at 18 Mbps.[5]
Encoding methodsCountless methods for encoding data in DNA are possible. The optimal methods are those that make economical use of DNA and protect against errors.[6] If the message DNA is intended to be stored for a long period of time, for example, 1,000 years [–> a lot longer than most of our digital storage media will likely last], it is also helpful if the sequence is obviously artificial and the reading frame is easy to identify.[6]
CNet gives details:
the next storage technology might use an approach as old as life on earth: DNA. Startup Catalog announced Friday it’s crammed all of the text of Wikipedia’s English-language version onto the same genetic molecules our own bodies use.
It accomplished the feat with its first DNA writer, a machine that would fit easily in your house if you first got rid of your refrigerator, oven and some counter space. And although it’s not likely to push aside your phone’s flash memory chips anytime soon, the company believes it’s useful already to some customers who need to archive data.
DNA strands are tiny and tricky to manage, but the biological molecules can store other data than the genes that govern how a cell becomes a pea plant or chimpanzee. Catalog uses prefabricated synthetic DNA strands that are shorter than human DNA, but uses a lot more of them so it can store much more data.
Relying on DNA instead of the latest high-tech miniaturization might sound like a step backward. But DNA is compact, chemically stable — and given that it’s the foundation of the Earth’s biology, it’s arguably not as likely to become as obsolete as the spinning magnetized platters of hard drives or CDs that are disappearing today . . .
In short, they used a different encoding and have stored Wikipedia in DNA.
At this point, we need to ask, why is it that we have seen certain objectors from the penumbra of attack sites making strident, unyielding objections to understanding DNA and mRNA as string data structure information storage entities, part of a wider information processing, protein synthesis process in the cell?
The manifest answer is simple and sad: because such things point to design, which is being ideologically locked out at all costs.
So, it is time to recognise a key first fact about DNA and mRNA and let the chips lie where they fly. END
