 Winston Ewert, one of the authors of Introduction to Evolutionary Informatics has a new paper at BIO-Complexity:
Winston Ewert, one of the authors of Introduction to Evolutionary Informatics has a new paper at BIO-Complexity:
Abstract: The hierarchical classification of life has been claimed as compelling evidence for universal common ancestry. However, research has uncovered much data which is not congruent with the hierarchical pattern. Nevertheless, biological data resembles a nested hierarchy sufficiently well to require an explanation. While many defenders of intelligent design dispute common descent, no alternative account of the approximate nested hierarchy pattern has been widely adopted. We present the dependency graph hypothesis as an alternative explanation, based on the technique used by software developers to reuse code among different software projects. This hypothesis postulates that different biological species share modules related by a dependency graph. We evaluate several predictions made by this model about both biological and synthetic data, finding them to be fulfilled. (open access) More.

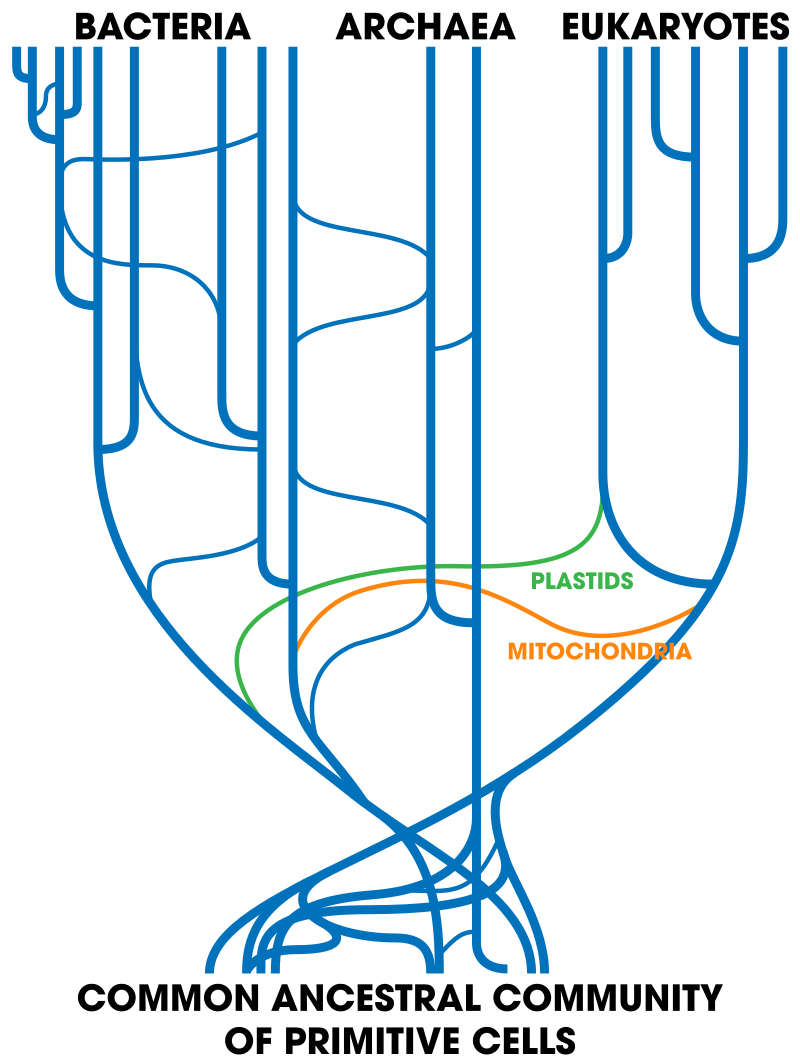
A friend offers a super-short summary:
1) Modularization is a well-established design methodology
2) Things that are developed with modularization can look as if they have a tree-like structure
3) A bayesian methodology can be used to differentiate between modularized and tree structures
4) For instances where the origins are known, the bayesian methodology correctly separates trees and modularizations
5) According to the methodology, genomes are organized more module-like than tree-like
Watch for Ewert’s work at Mind Matters Today as well, in links from here.
See also: Podcast: Winston Ewert on computer simulation of evolution (AVIDA) that sneaks in information
Dr. Ewertanswers some questions
Open Mike: Cornell OBI Conference Chapter Six – Ewert et all on the Tierra evolution program – Summary
Open Mike: Cornell OBI Conference—Chapter Three, Dembski, Ewert, and Marks on the true cost of a successful search
Also: Bill Dembski on the Evolutionary Informatics Lab – the one a Baylor dean tried to shut down
Evolutionary informatics has come a long way since a Baylor dean tried to shut down the lab
Introduction to Evolutionary Informatics: Media to get you started
Who thinks Introduction to Evolutionary Informatics should be on your summer reading list?
and
Evolutionary Informatics takes off